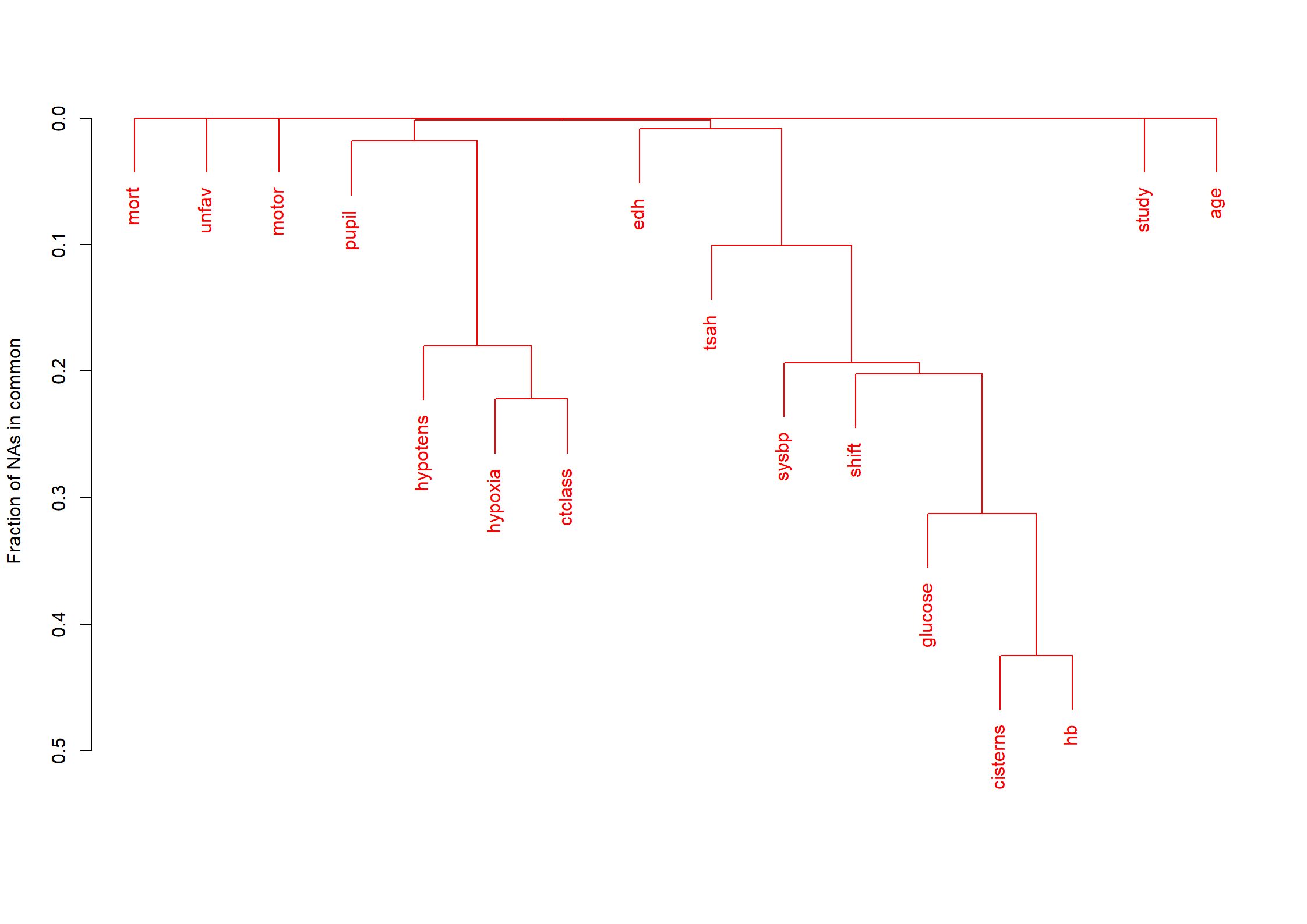
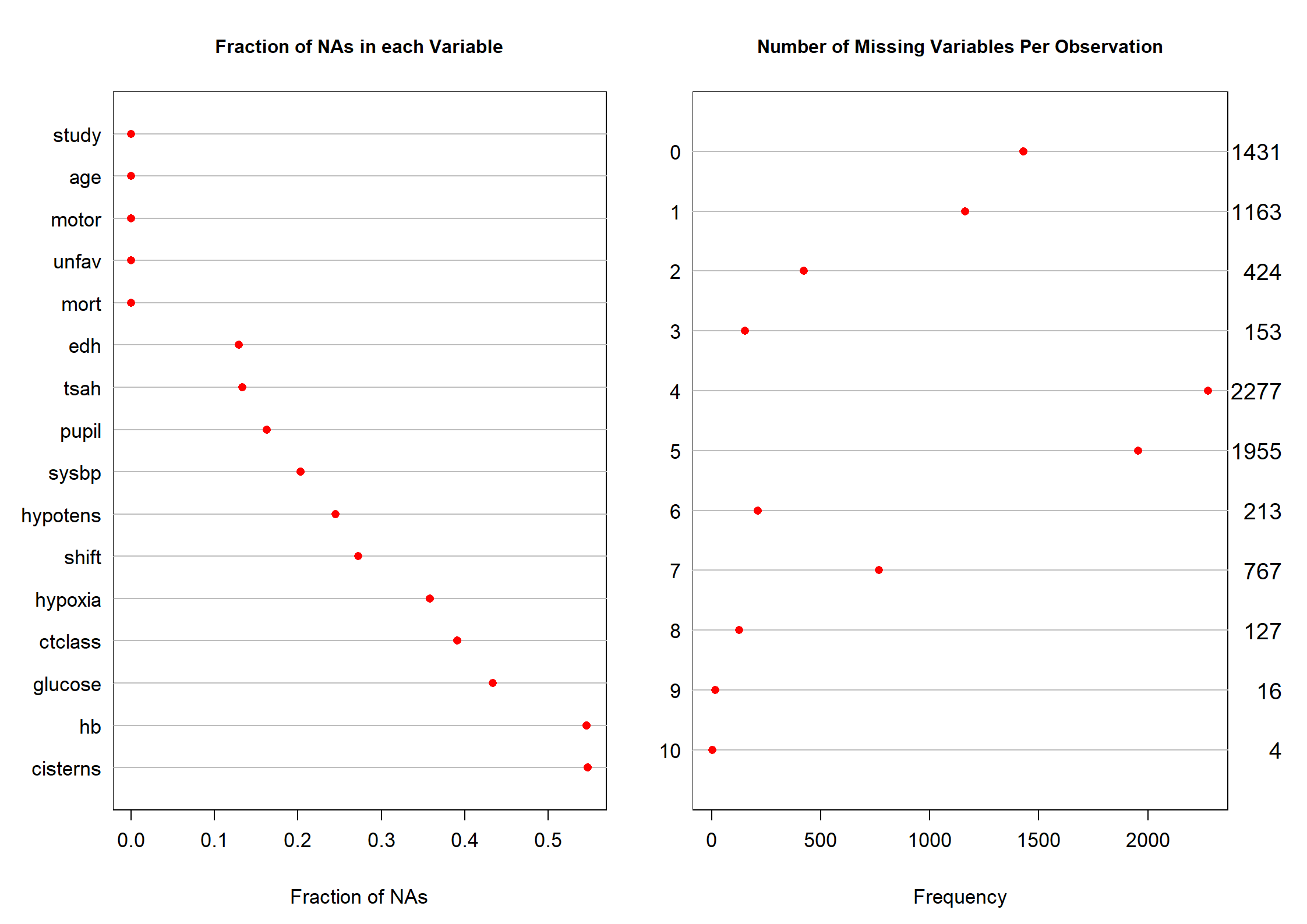
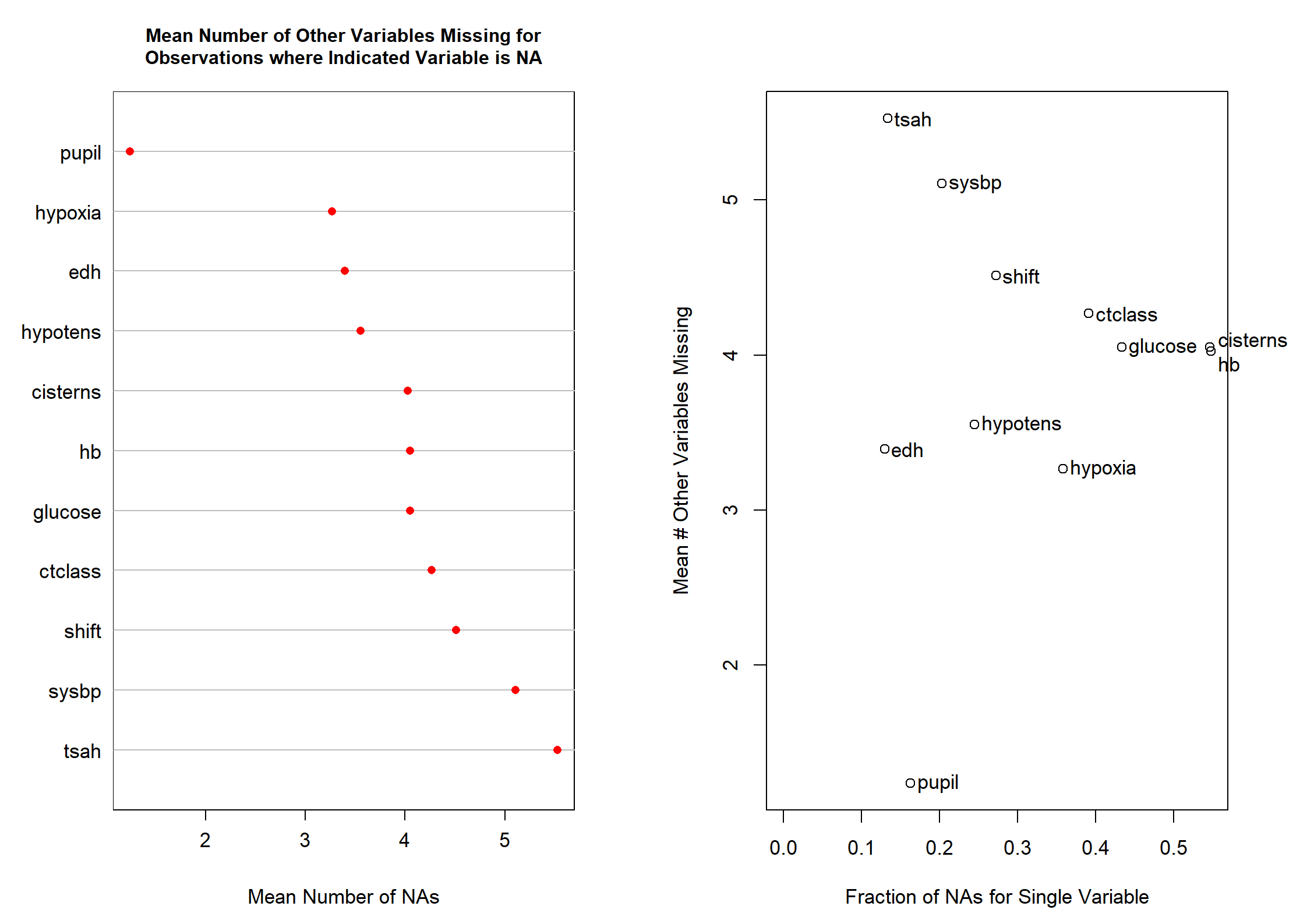
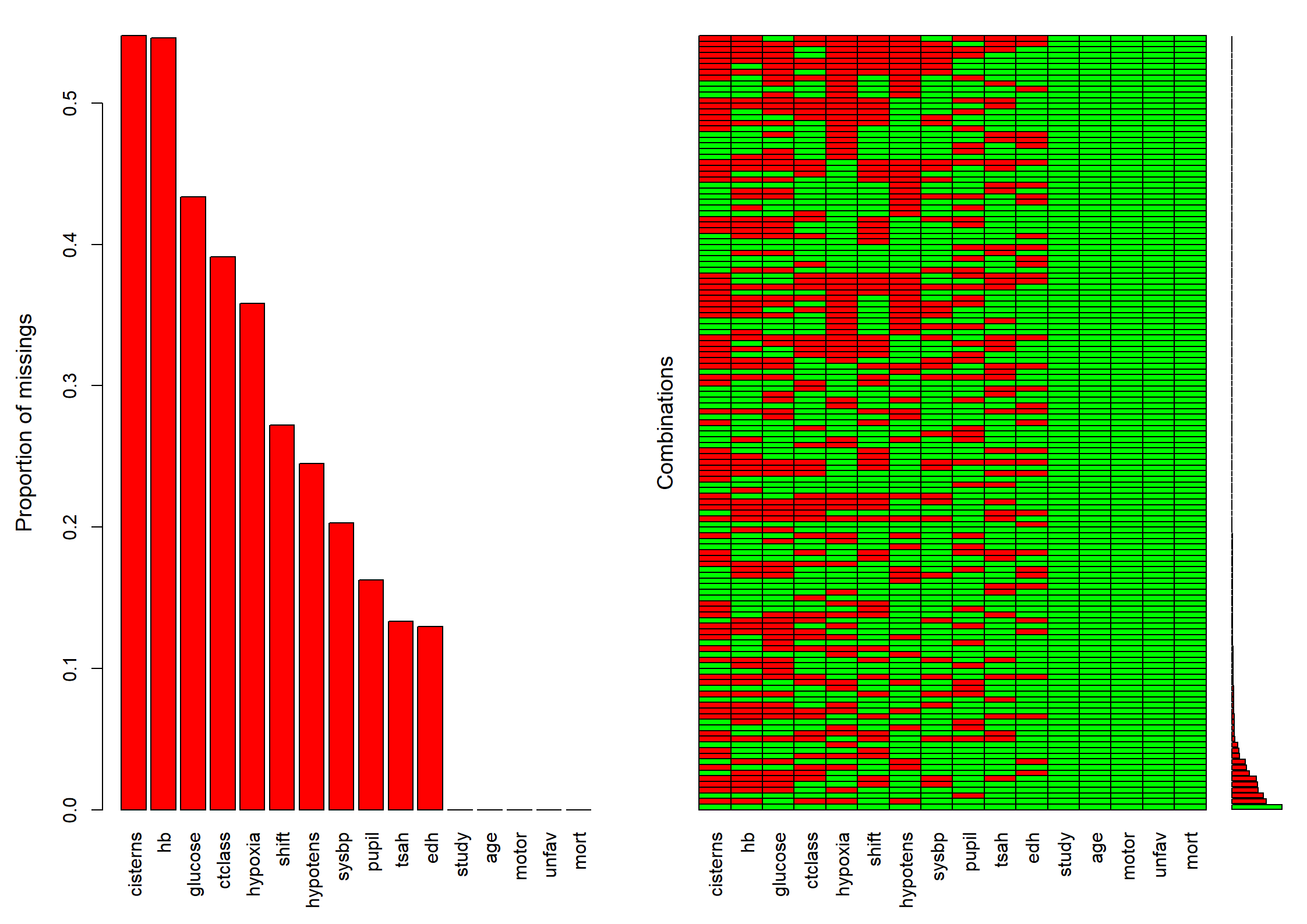
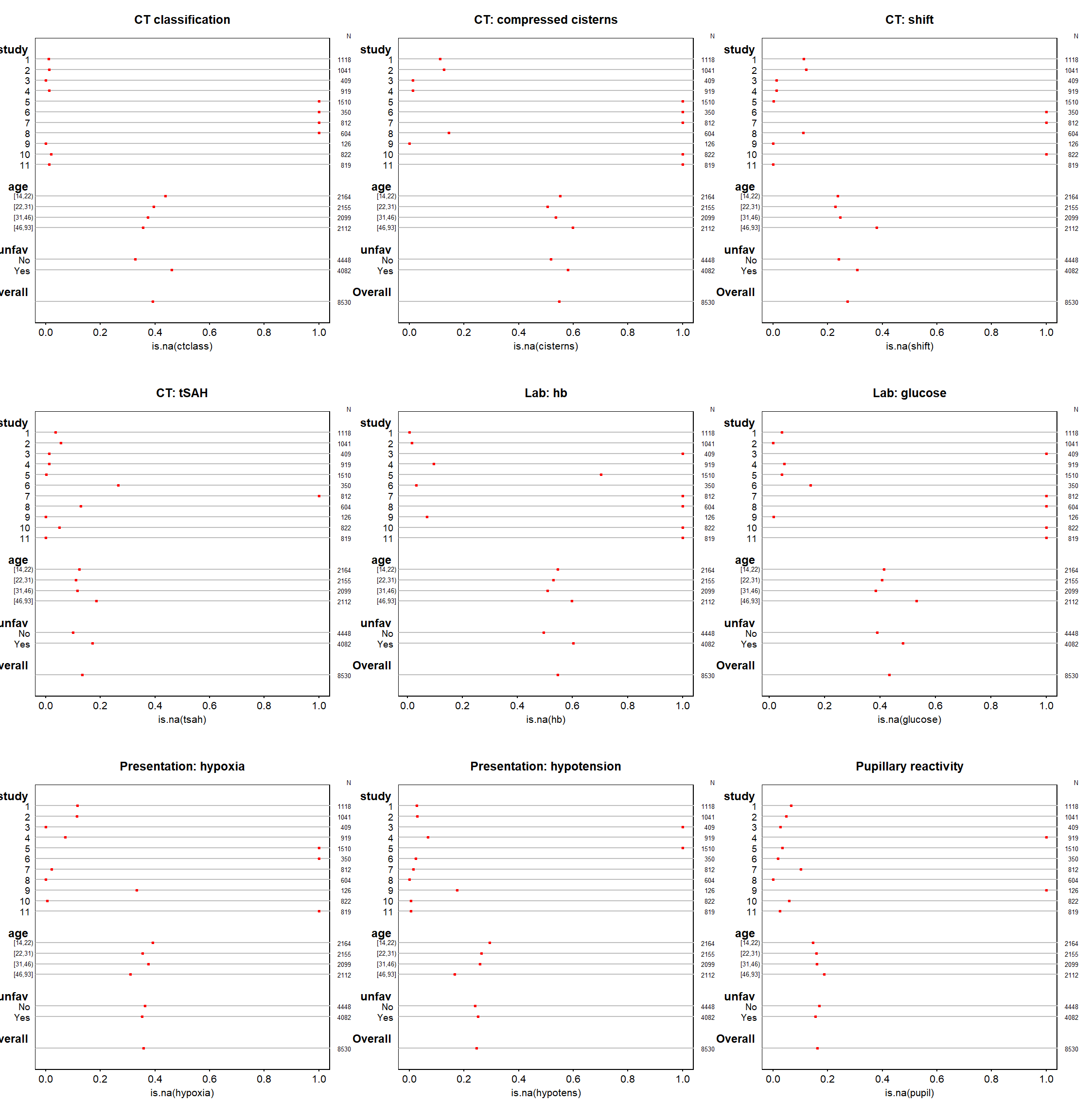
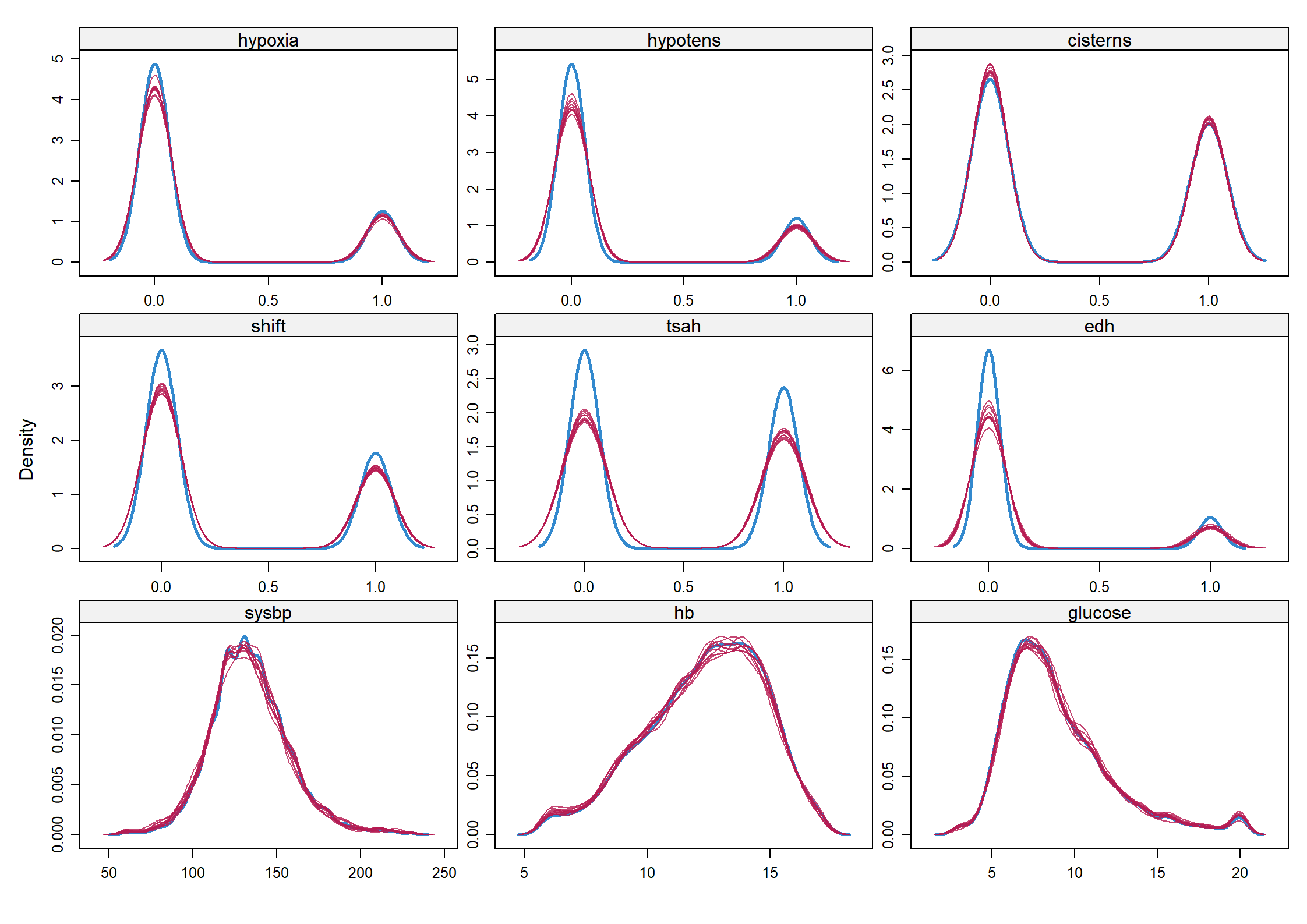
Code show/hide
TBI1 <- read.csv("data/TBI1.csv", row.names = 1)
TBI1$study <- as.factor(TBI1$study)
TBI1$pupil <- as.factor(TBI1$pupil)
TBI1$ctclass <- as.factor(TBI1$ctclass)
describe(TBI1)TBI1
16 Variables 8530 Observations
----------------------------------------------------------------------------------------------------
study
n missing distinct
8530 0 11
Value 1 2 3 4 5 6 7 8 9 10 11
Frequency 1118 1041 409 919 1510 350 812 604 126 822 819
Proportion 0.131 0.122 0.048 0.108 0.177 0.041 0.095 0.071 0.015 0.096 0.096
----------------------------------------------------------------------------------------------------
age
n missing distinct Info Mean Gmd .05 .10 .25 .50 .75
8530 0 80 0.999 34.7 17.59 17 18 21 30 45
.90 .95
59 65
lowest : 14 15 16 17 18, highest: 89 90 91 92 93
----------------------------------------------------------------------------------------------------
hypoxia
n missing distinct Info Sum Mean Gmd
5473 3057 2 0.489 1122 0.205 0.326
----------------------------------------------------------------------------------------------------
hypotens
n missing distinct Info Sum Mean Gmd
6440 2090 2 0.447 1173 0.1821 0.298
----------------------------------------------------------------------------------------------------
cisterns
n missing distinct Info Sum Mean Gmd
3857 4673 2 0.736 1661 0.4306 0.4905
----------------------------------------------------------------------------------------------------
shift
n missing distinct Info Sum Mean Gmd
6209 2321 2 0.658 2020 0.3253 0.4391
----------------------------------------------------------------------------------------------------
tsah
n missing distinct Info Sum Mean Gmd
7393 1137 2 0.742 3313 0.4481 0.4947
----------------------------------------------------------------------------------------------------
edh
n missing distinct Info Sum Mean Gmd
7425 1105 2 0.35 1001 0.1348 0.2333
----------------------------------------------------------------------------------------------------
pupil
n missing distinct
7143 1387 3
Value 1 2 3
Frequency 4498 887 1758
Proportion 0.630 0.124 0.246
----------------------------------------------------------------------------------------------------
motor
n missing distinct
8530 0 5
Value 1/2 3 4 5/6 9
Frequency 2439 1085 1941 2602 463
Proportion 0.286 0.127 0.228 0.305 0.054
----------------------------------------------------------------------------------------------------
ctclass
n missing distinct
5192 3338 3
Value 1 2 3
Frequency 2198 1050 1944
Proportion 0.423 0.202 0.374
----------------------------------------------------------------------------------------------------
sysbp
n missing distinct Info Mean Gmd .05 .10 .25 .50 .75
6797 1733 1653 0.999 134.3 25.3 100 108 120 132 148
.90 .95
162 175
lowest : 60 62 64 64.19 65.6 , highest: 219 220 222 226 230
----------------------------------------------------------------------------------------------------
hb
n missing distinct Info Mean Gmd .05 .10 .25 .50 .75
3871 4659 195 1 12.39 2.685 8.1 9.1 10.8 12.6 14.2
.90 .95
15.2 15.8
lowest : 6 6.1 6.12311 6.2 6.28424, highest: 16.7 16.79 16.8 16.9 17
----------------------------------------------------------------------------------------------------
glucose
n missing distinct Info Mean Gmd .05 .10 .25 .50 .75
4830 3700 706 1 8.894 3.393 5.100 5.611 6.700 8.200 10.400
.90 .95
13.091 15.500
lowest : 3 3.03 3.05556 3.2 3.3 , highest: 19.7778 19.8 19.855 19.9 20
----------------------------------------------------------------------------------------------------
unfav
n missing distinct Info Sum Mean Gmd
8530 0 2 0.749 4082 0.4785 0.4991
----------------------------------------------------------------------------------------------------
mort
n missing distinct Info Sum Mean Gmd
8530 0 2 0.606 2396 0.2809 0.404
----------------------------------------------------------------------------------------------------