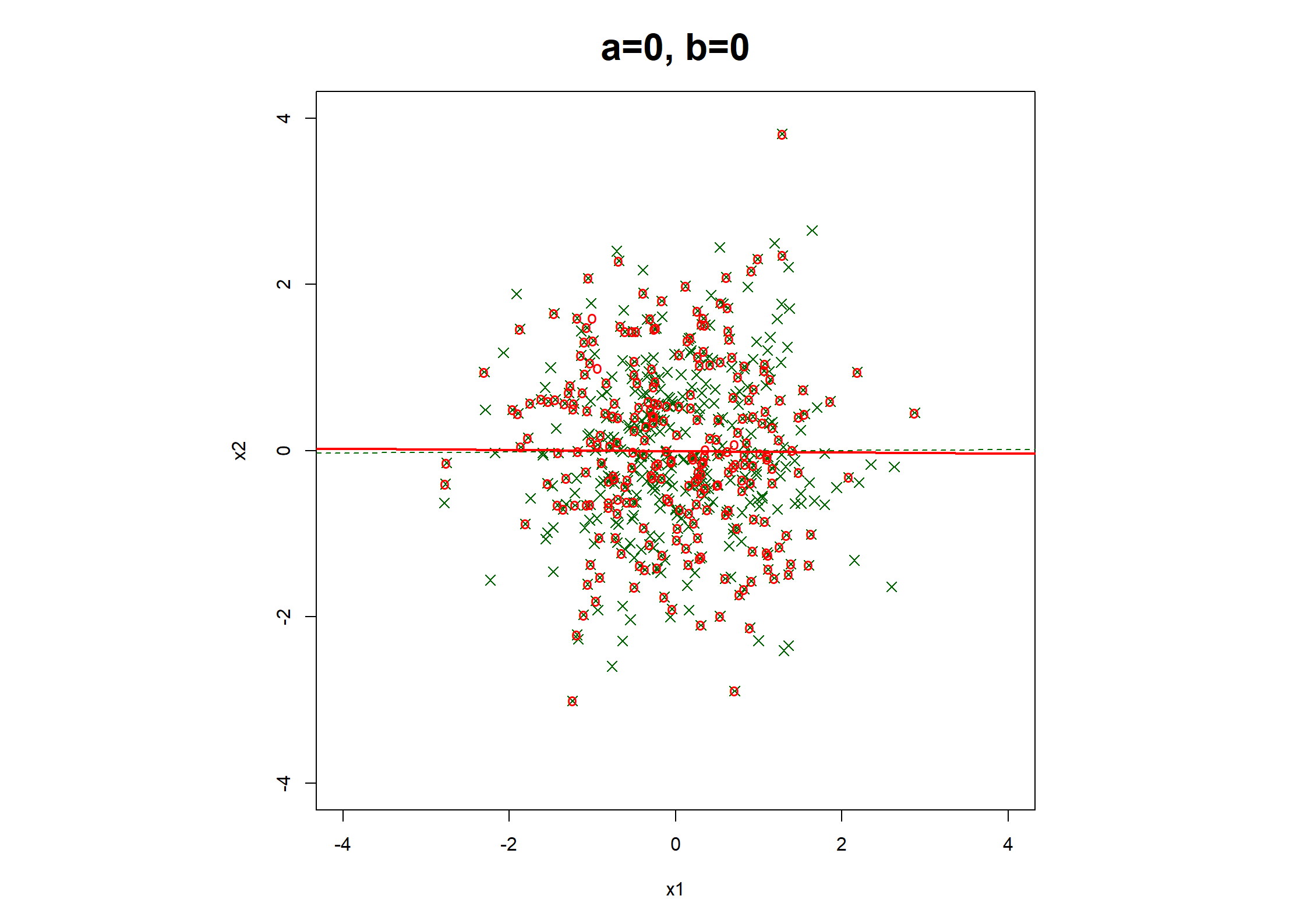
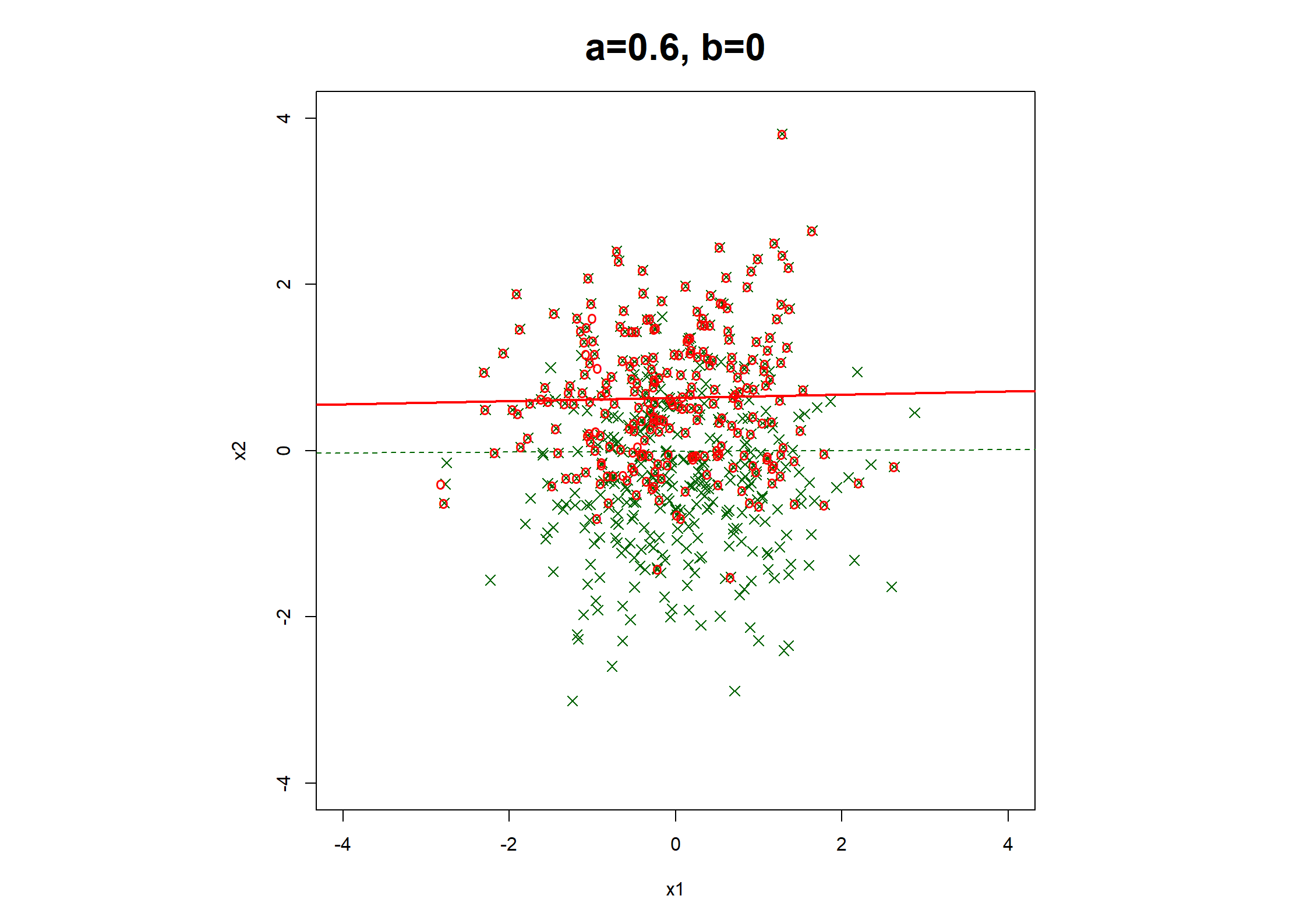
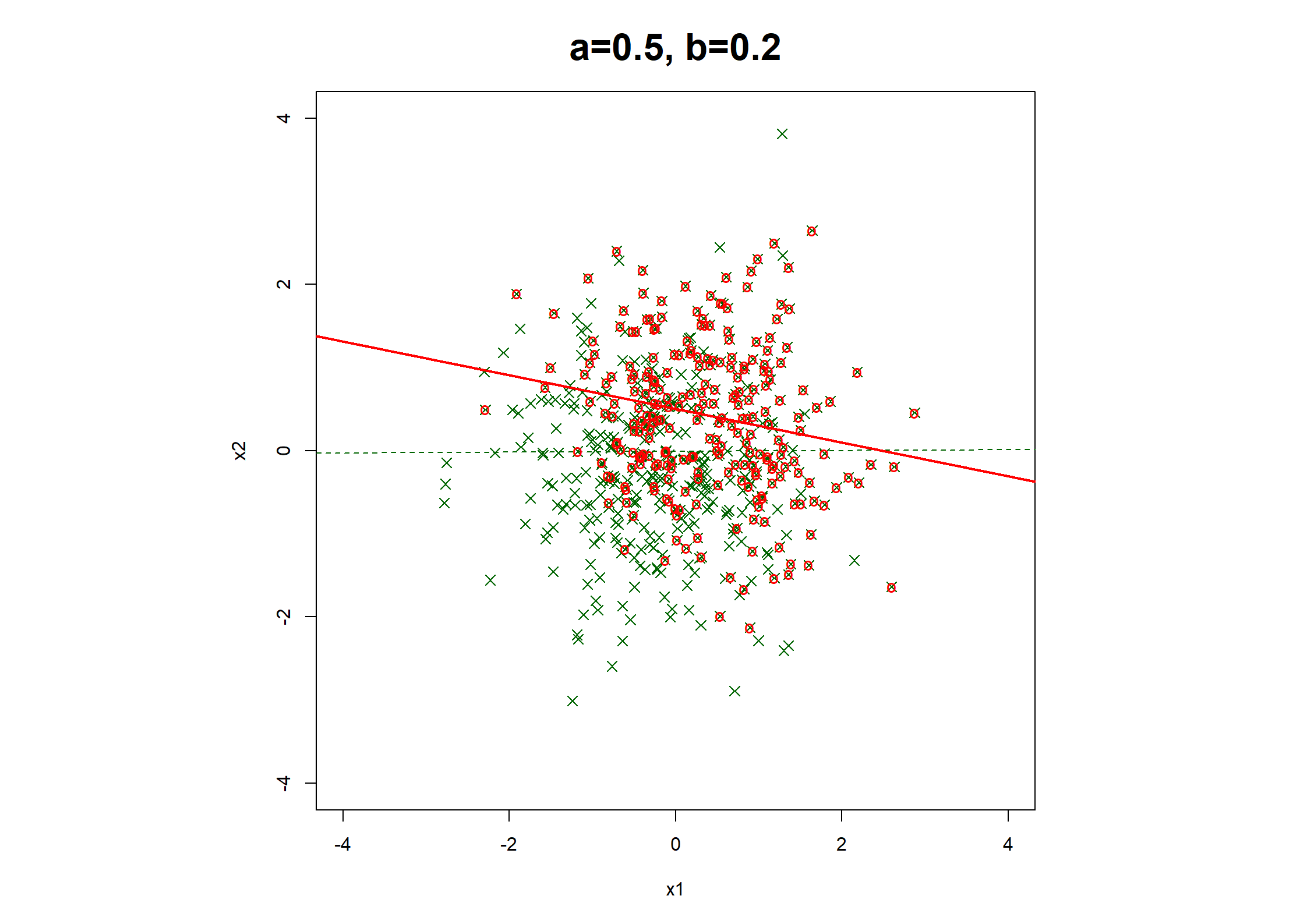
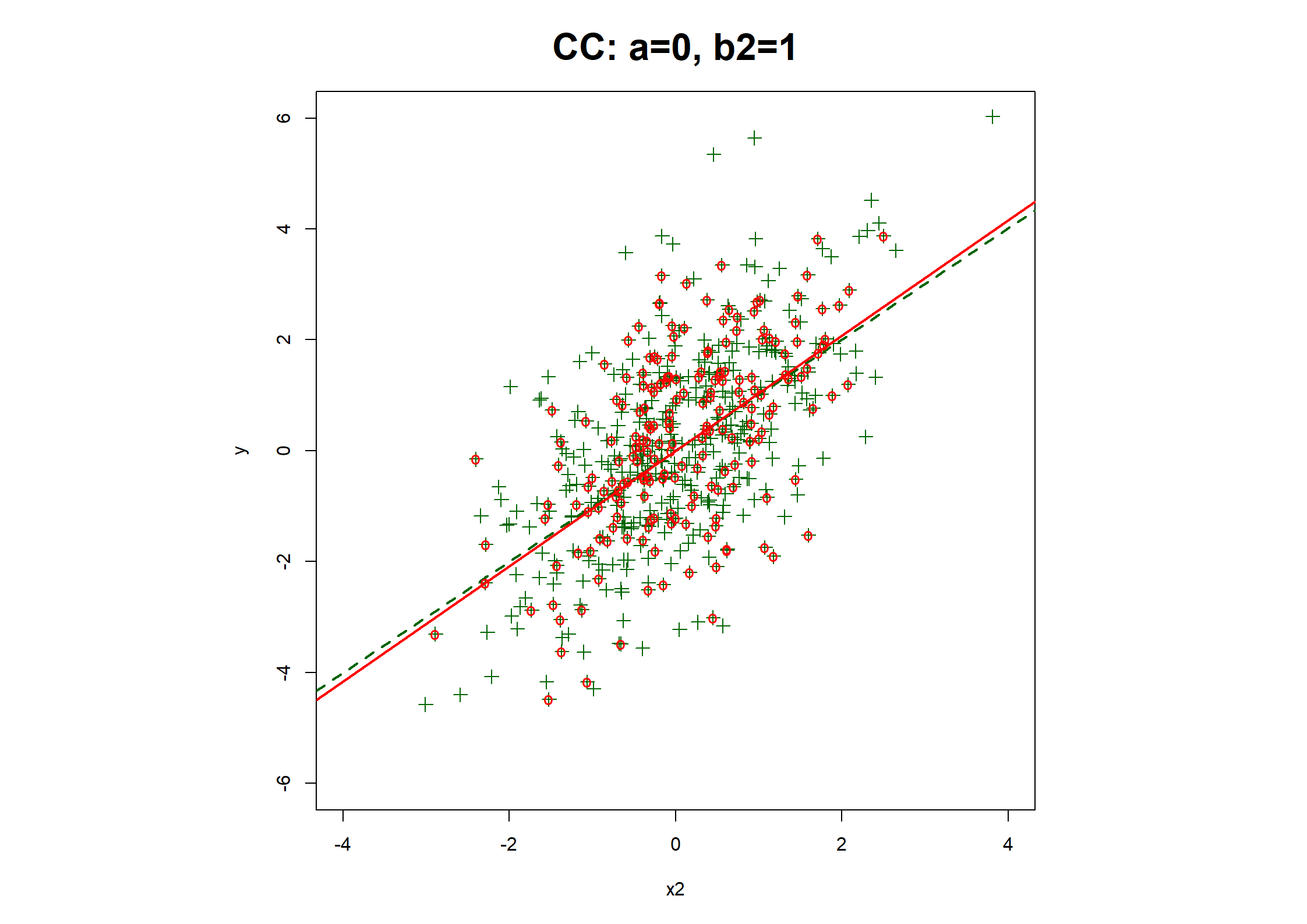
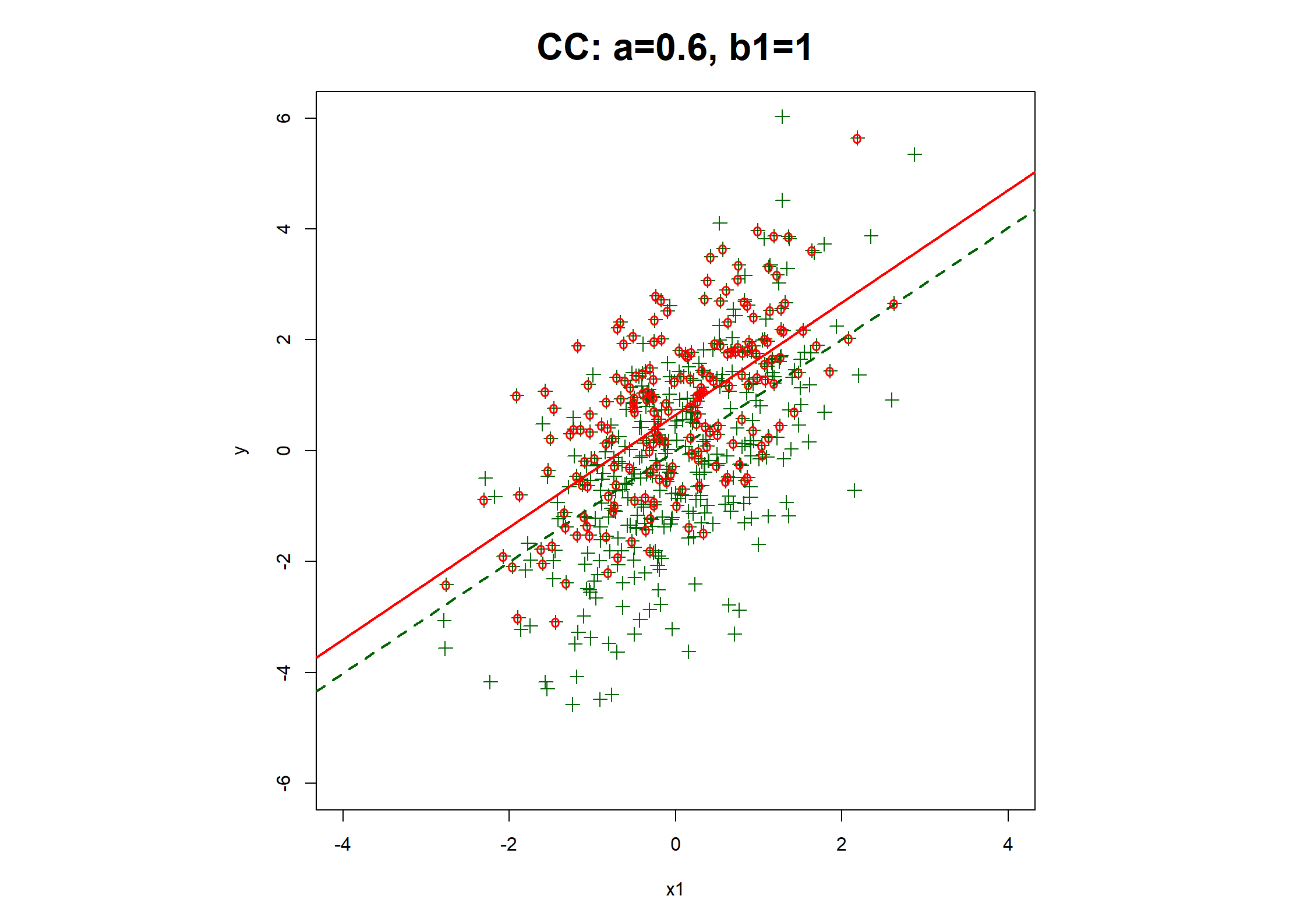
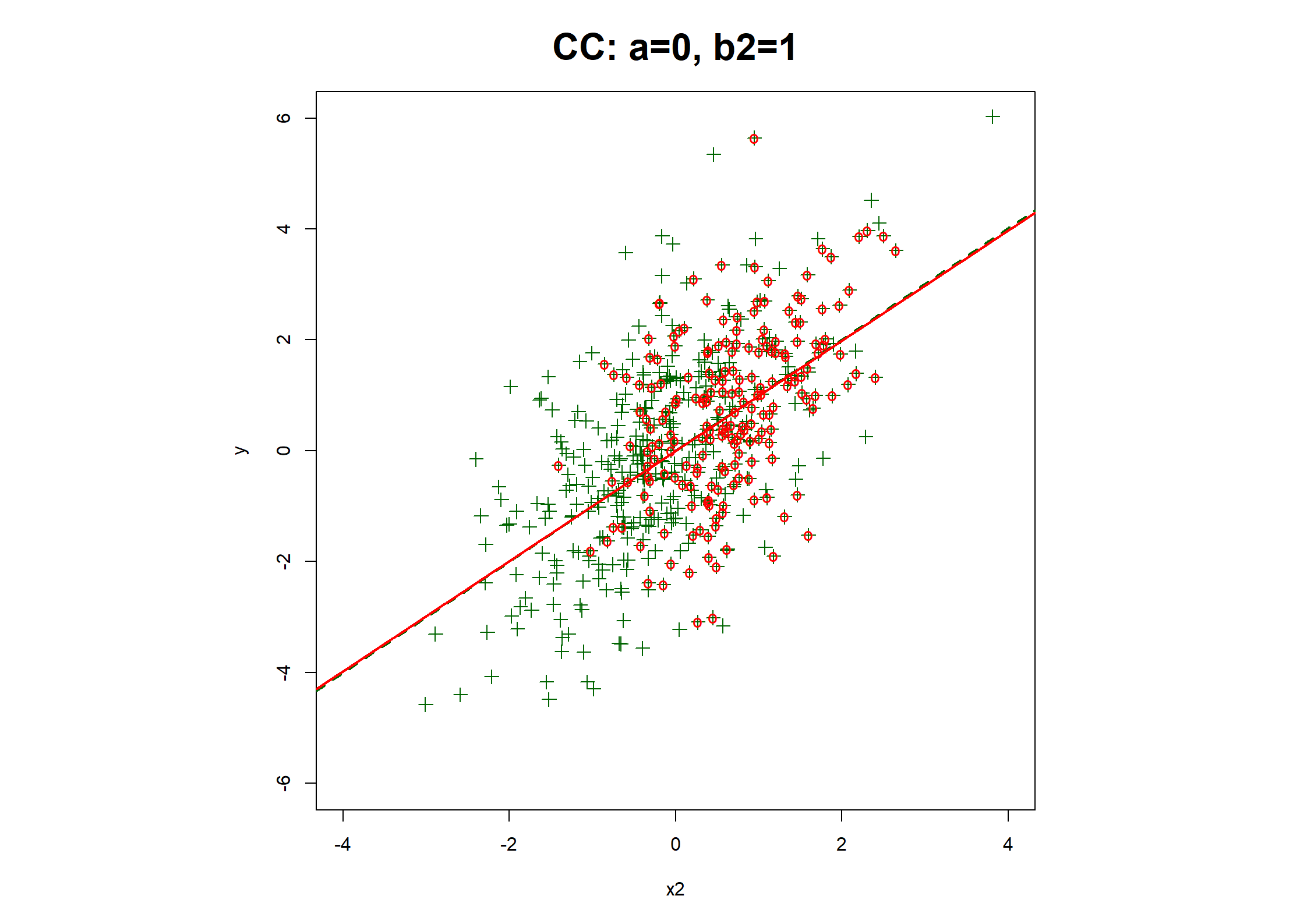
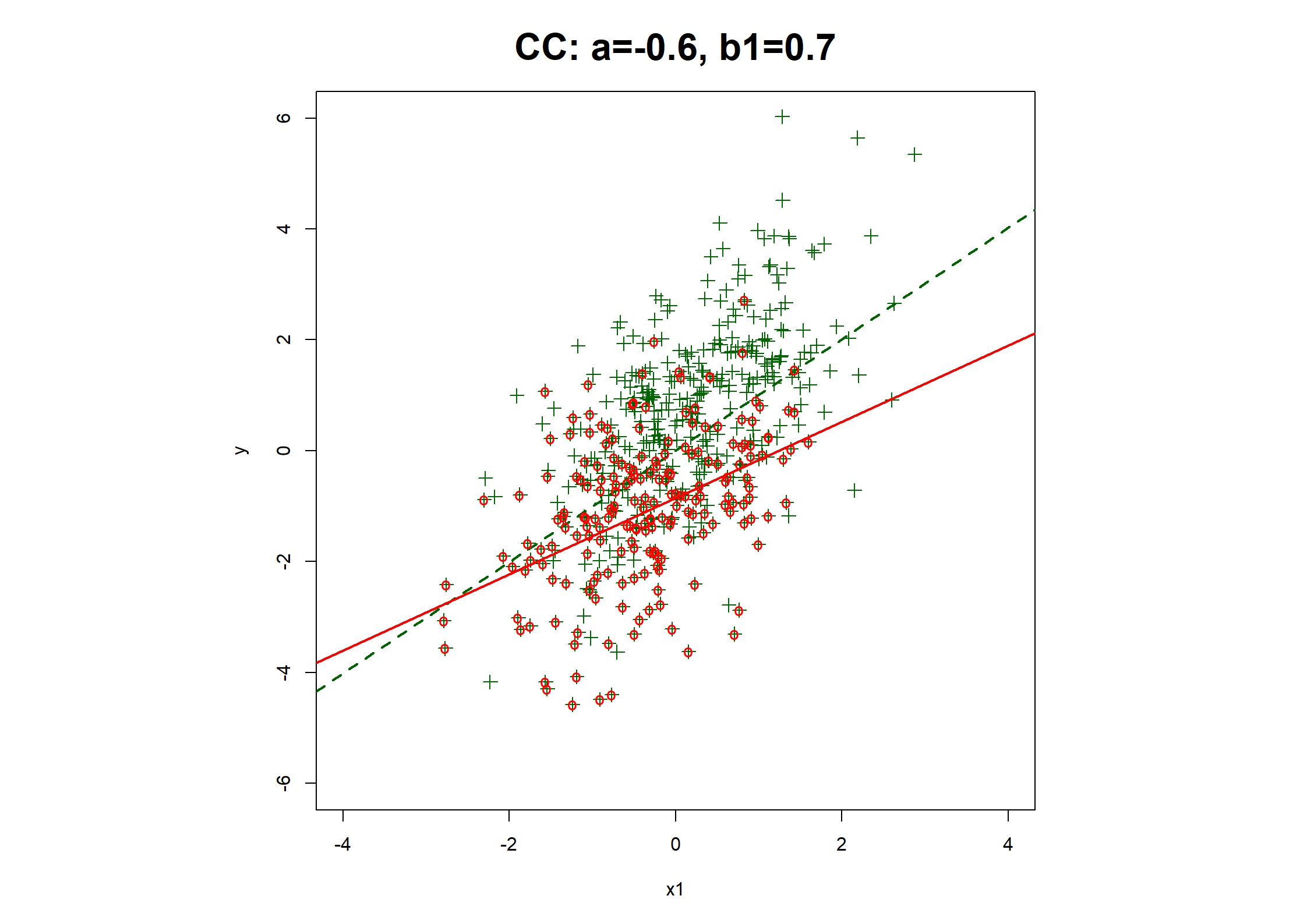
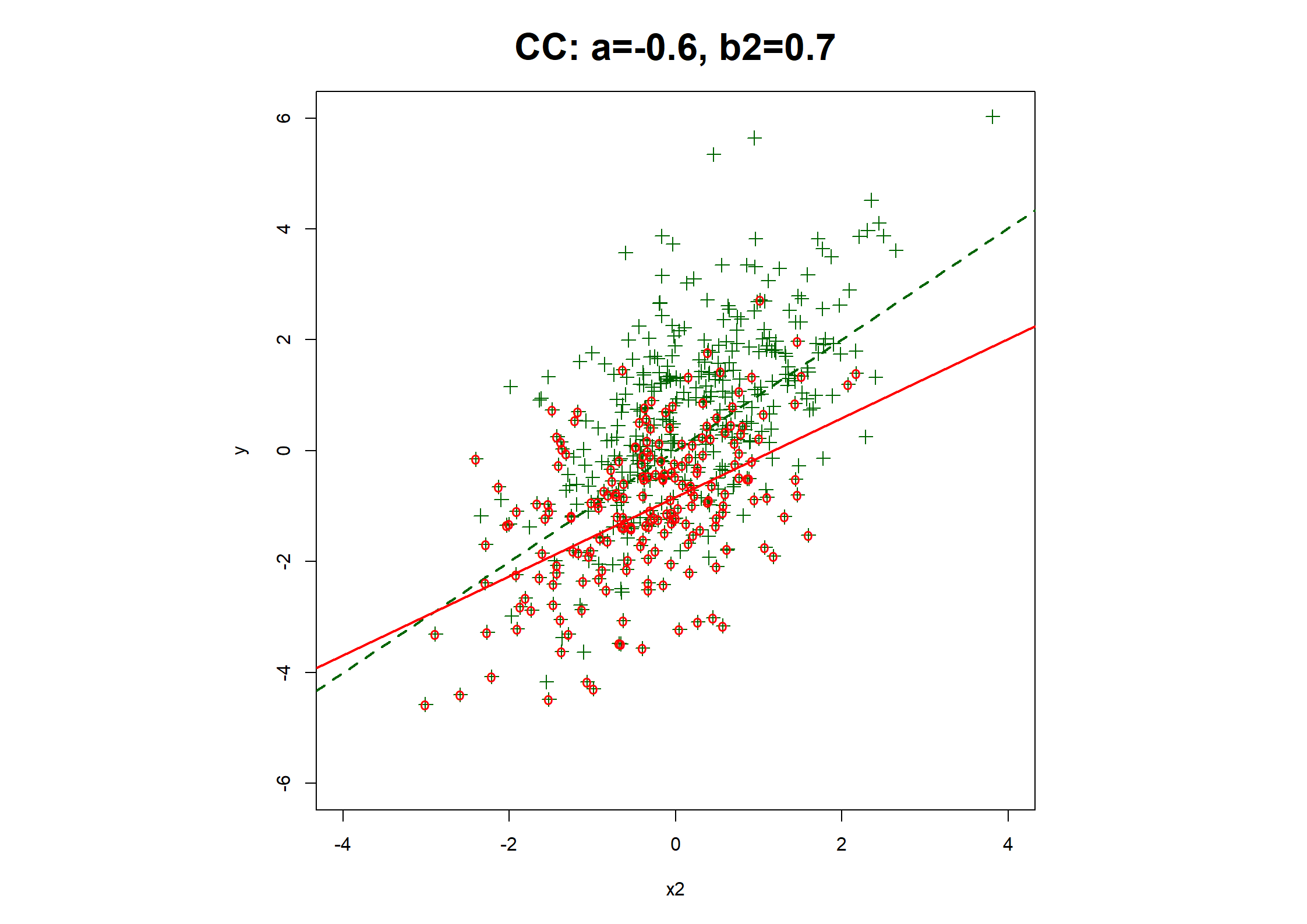
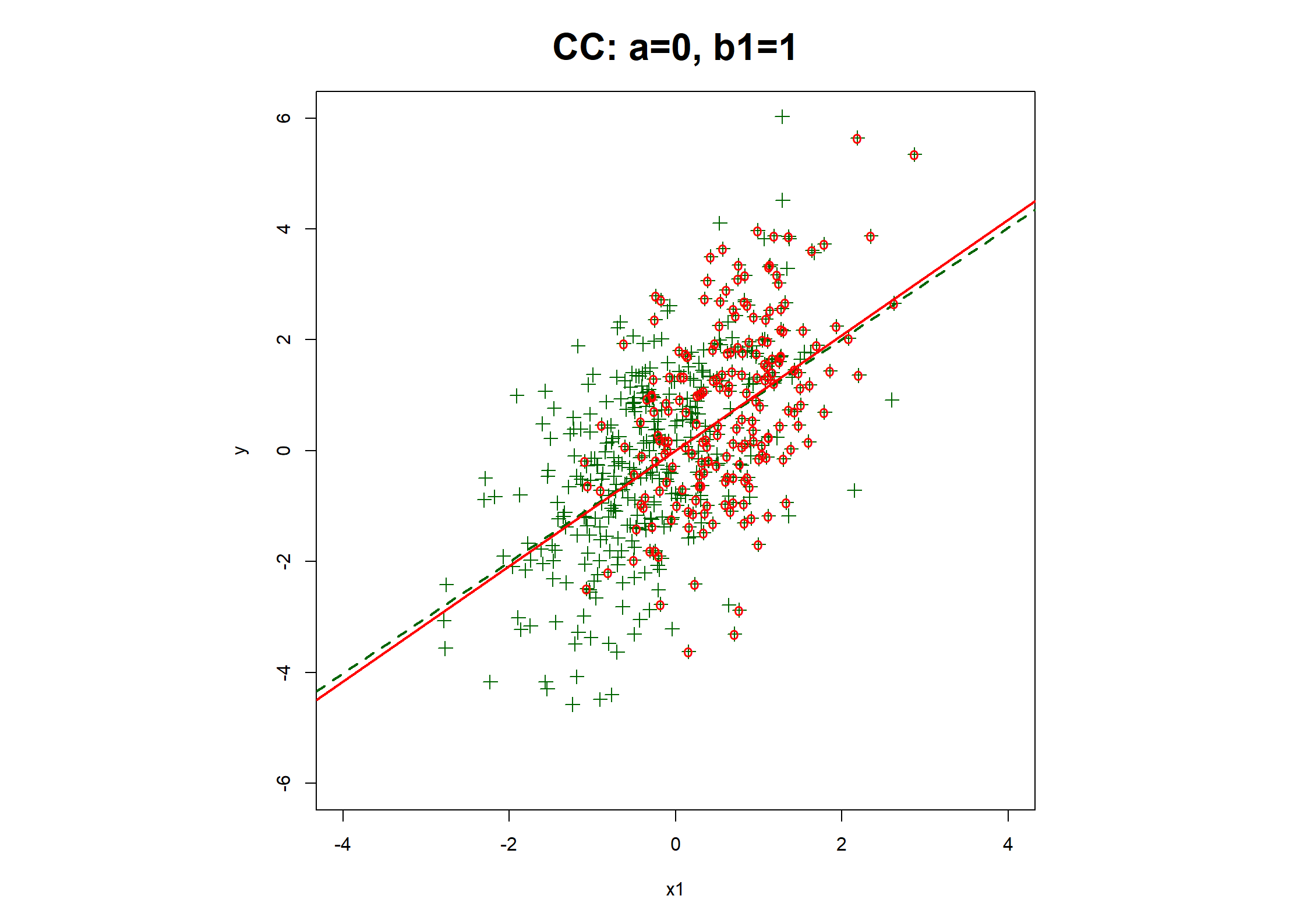
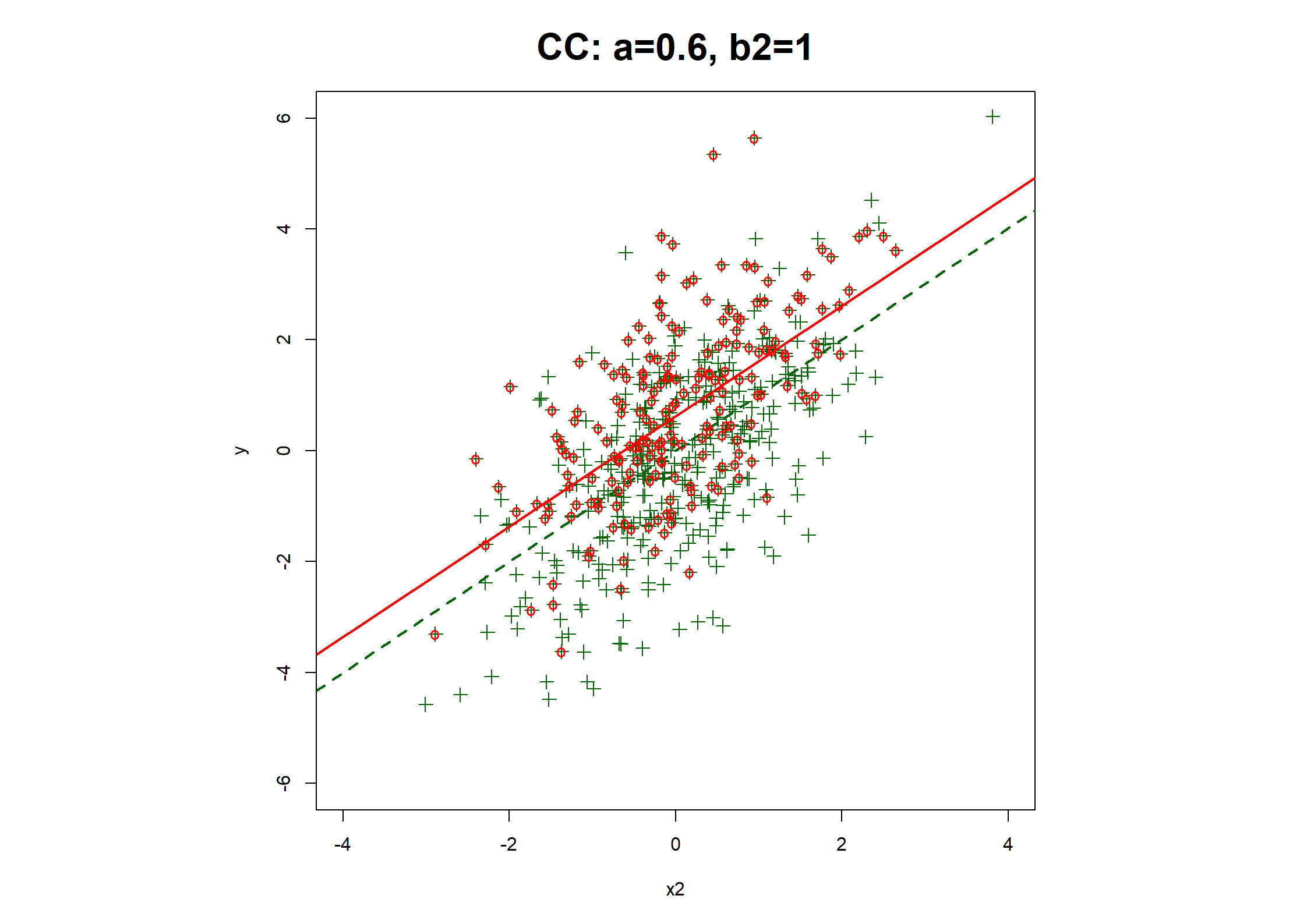
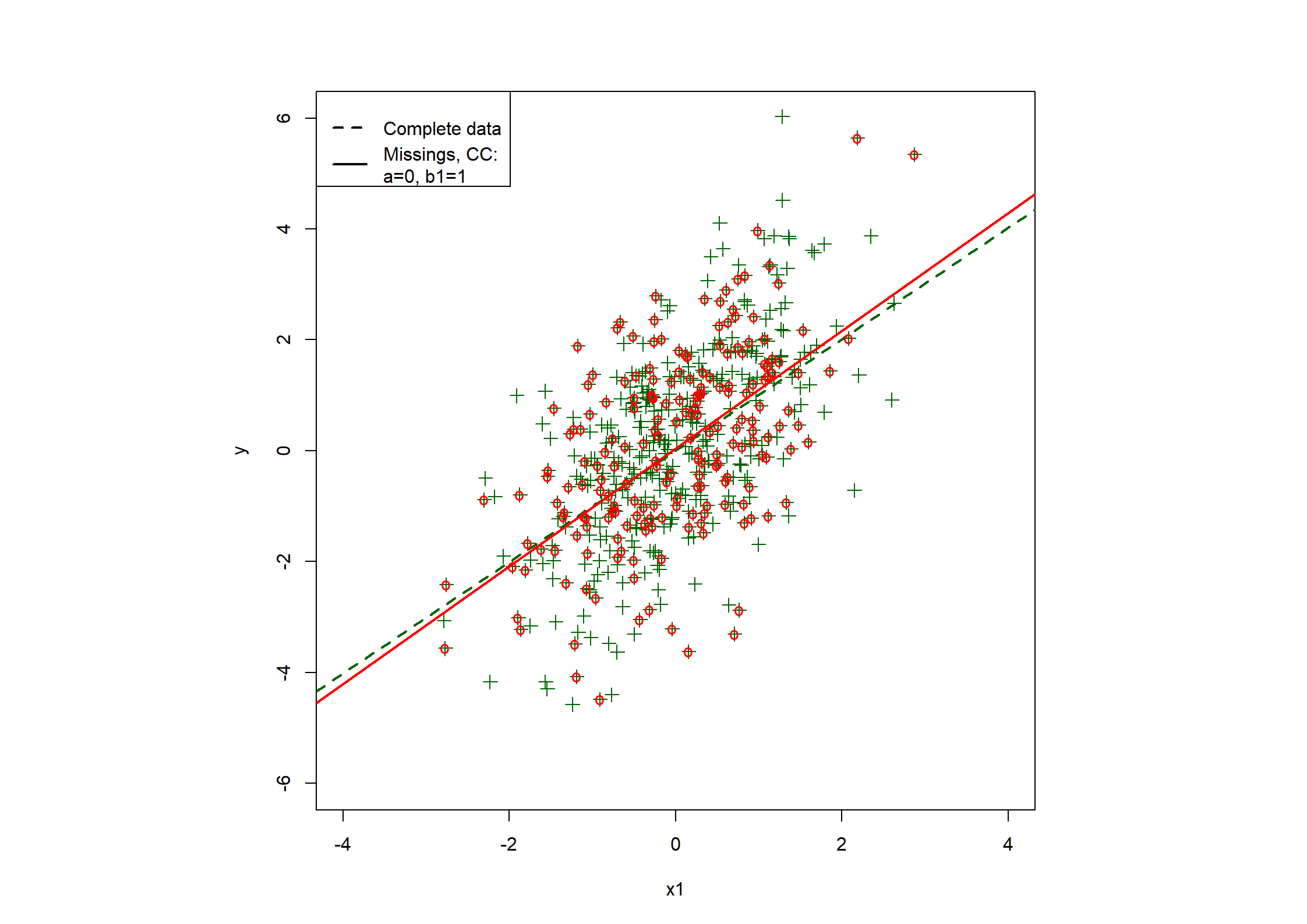
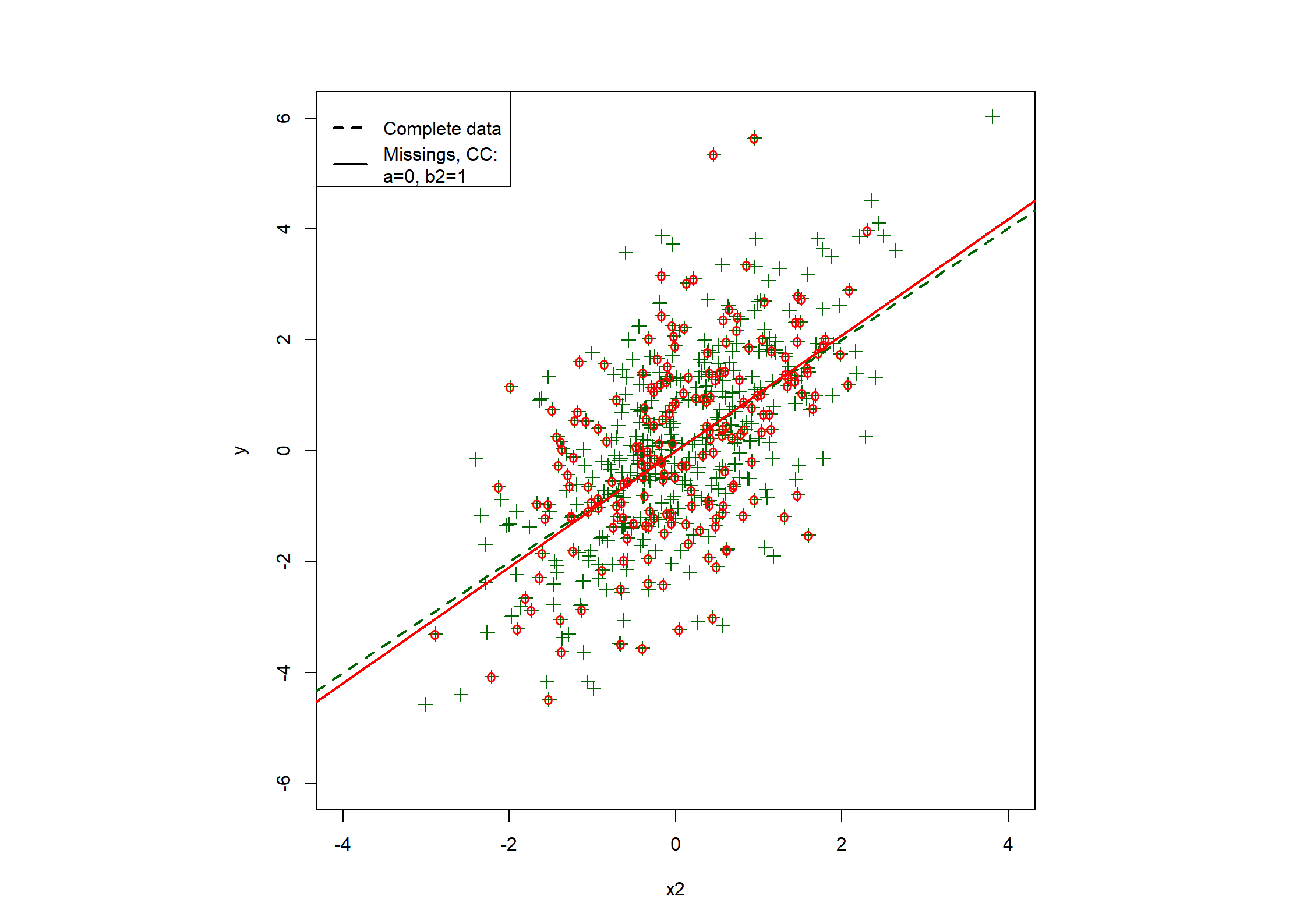
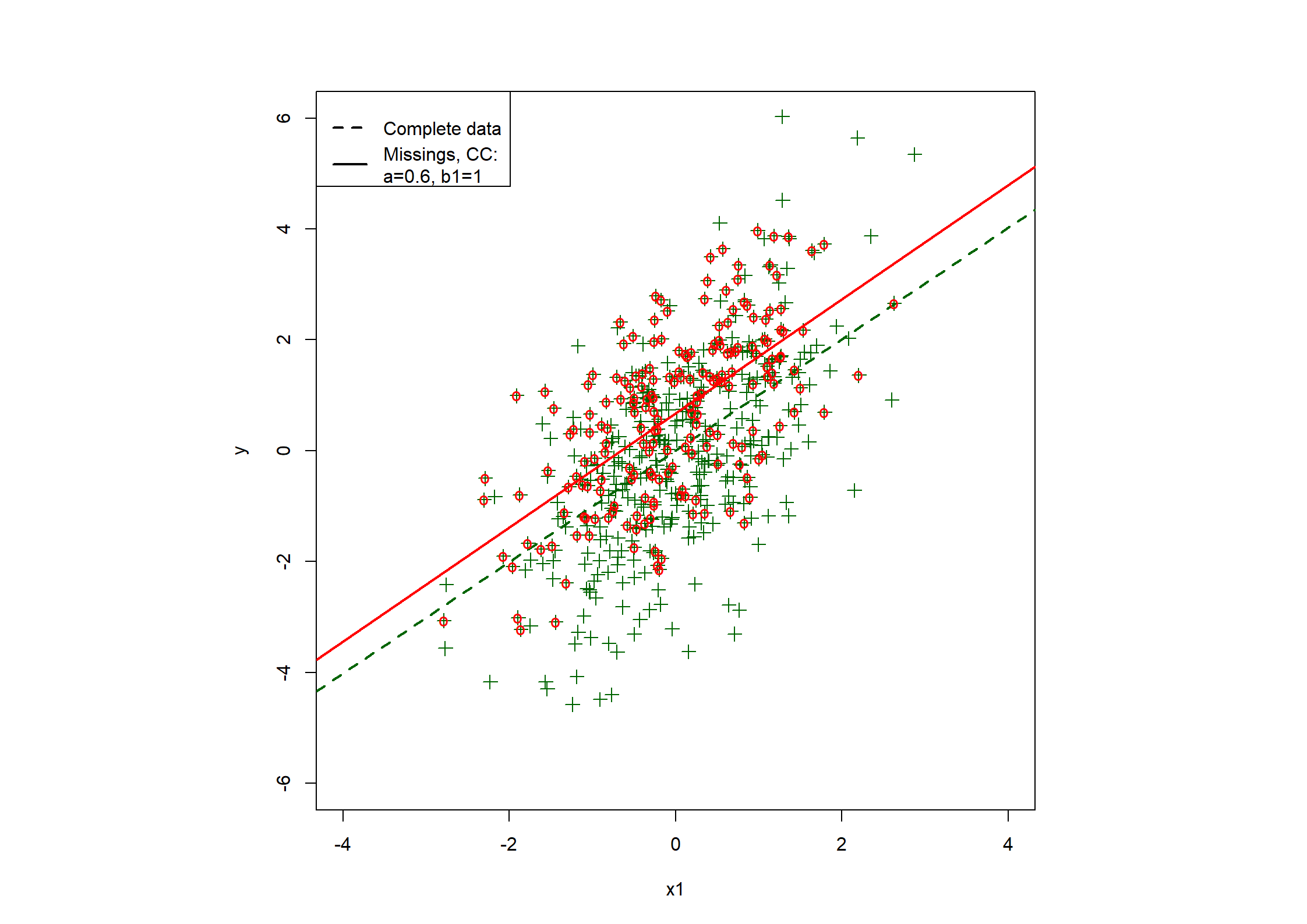
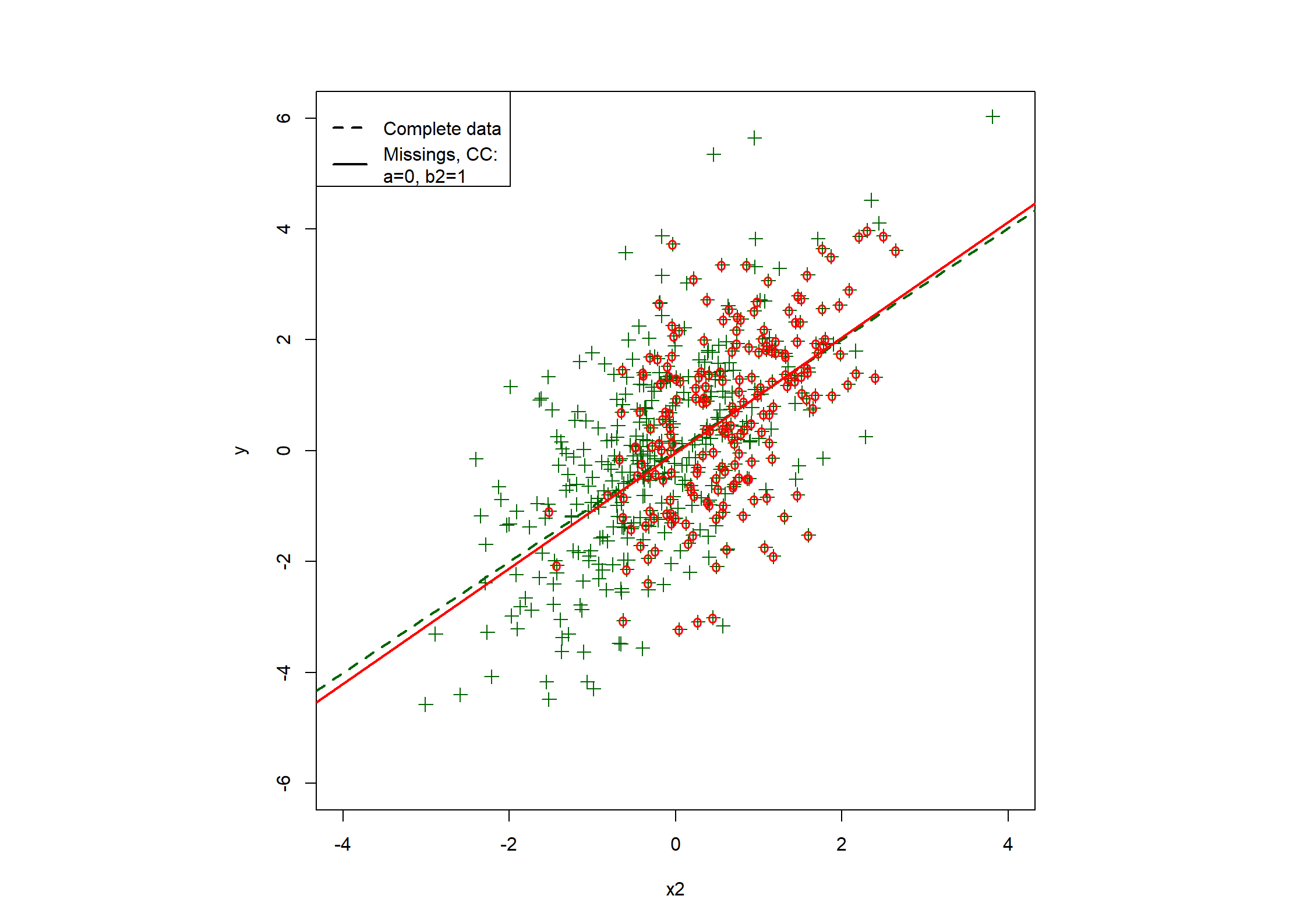
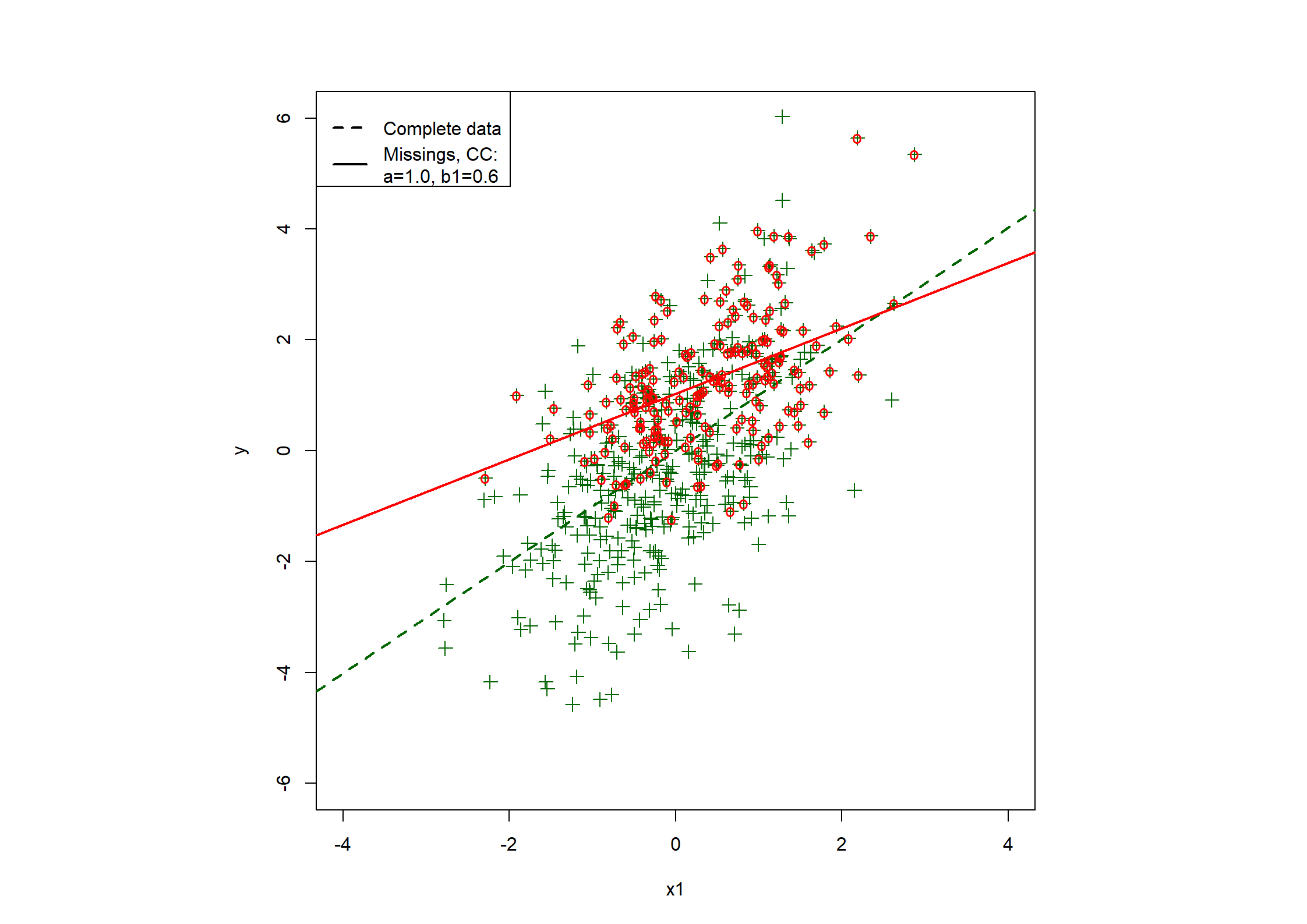
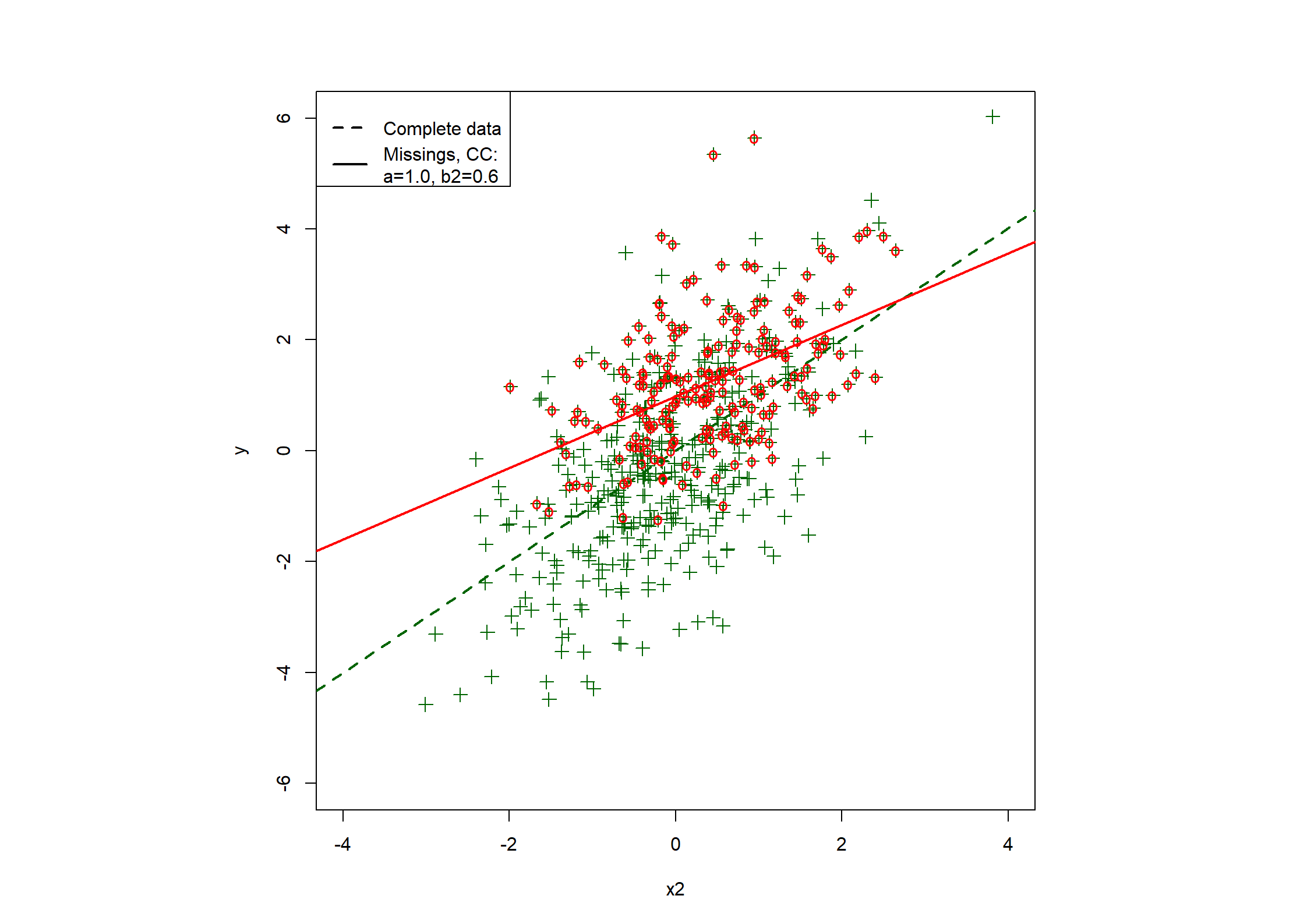
## Table 7.6 ##
# Regression coefficients and R2
print(rbind(
c(coef(fx1.CC), fx1.CC$stats["R2"]),
c(coef(fx1MCAR.CC), fx1MCAR.CC$stats["R2"]),
c(coef(fx1MARx.CC), fx1MARx.CC$stats["R2"]),
c(coef(fx1MARy.CC), fx1MARy.CC$stats["R2"]),
c(coef(fx1MNAR.CC), fx1MNAR.CC$stats["R2"]),
c(coef(fy.CC), fy.CC$stats["R2"]),
c(coef(fyMARx.CC), fyMARx.CC$stats["R2"]),
c(coef(fyMNAR.CC), fyMNAR.CC$stats["R2"])
),
digits = 2
)
# Imputation; make data sets with different types of missings
d <- as.data.frame(cbind(y1, x1, x2, x1MCAR, x1MARx, x1MARy, x1MNAR))
d <- d[1:500000, ]
dMCAR <- d[, c(1, 3, 4)] # data set with x1 missings according to MCAR
dMARx <- d[, c(1, 3, 5)] # x1 MAR on x2
dMARy <- d[, c(1, 3, 6)] # x1 MAR on y
dMNAR <- d[, c(1, 3, 7)] # x1 MNAR at x1
dy <- as.data.frame(cbind(y1, x1, x2, yMCAR, yMARx2, yMNAR))
dy <- dy[1:500000, ]
dyMCAR <- d[, c(2, 3, 4)] # data set with y missing MCAR
dyMARx <- d[, c(2, 3, 5)] # x1 MAR on x2
dyMNAR <- d[, c(2, 3, 6)] # x1 MAR on x2
## Imputation using the aregImpute function from rms
m <- 1 # to be changed
g <- aregImpute(~ y1 + x1MCAR + x2, n.impute = m, data = d, pr = F, type = "pmm")
## fit models per imputed set and combine results using Rubin's rules
## Use fit.mult.impute() function from rms
fx1MCAR.MI.u <- fit.mult.impute(y1 ~ x1MCAR, ols, xtrans = g, data = d, pr = F)
fx2MCAR.MI.u <- fit.mult.impute(y1 ~ x2, ols, xtrans = g, data = d, pr = F)
fx1MCAR.MI <- fit.mult.impute(y1 ~ x1MCAR + x2, ols, xtrans = g, data = d, pr = F)
g <- aregImpute(~ y1 + x1MARx + x2, n.impute = m, data = d, pr = F, type = "pmm")
fx1MARx.MI.u <- fit.mult.impute(y1 ~ x1MARx, ols, xtrans = g, data = d, pr = F)
fx2MARx.MI.u <- fit.mult.impute(y1 ~ x2, ols, xtrans = g, data = d, pr = F)
fx1MARx.MI <- fit.mult.impute(y1 ~ x1MARx + x2, ols, xtrans = g, data = d, pr = F) ## areg
g <- aregImpute(~ y1 + x1MARy + x2, n.impute = m, data = d, pr = F, type = "pmm")
fx1MARy.MI.u <- fit.mult.impute(y1 ~ x1MARy, ols, xtrans = g, data = d, pr = F)
fx2MARy.MI.u <- fit.mult.impute(y1 ~ x2, ols, xtrans = g, data = d, pr = F)
fx1MARy.MI <- fit.mult.impute(y1 ~ x1MARy + x2, ols, xtrans = g, data = d, pr = F) ## areg
g <- aregImpute(~ y1 + x1MNAR + x2, n.impute = m, data = d, pr = F, type = "pmm")
fx1MNAR.MI.u <- fit.mult.impute(y1 ~ x1MNAR, ols, xtrans = g, data = d, pr = F)
fx2MNAR.MI.u <- fit.mult.impute(y1 ~ x2, ols, xtrans = g, data = d, pr = F)
fx1MNAR.MI <- fit.mult.impute(y1 ~ x1MNAR + x2, ols, xtrans = g, data = d, pr = F) ## areg
# Regression coefficients after MI with aregImpute
print(rbind(
coef(fx1MCAR.MI.u), coef(fx2MCAR.MI.u), coef(fx1MCAR.MI),
coef(fx1MARx.MI.u), coef(fx2MARx.MI.u), coef(fx1MARx.MI),
coef(fx1MARy.MI.u), coef(fx2MARy.MI.u), coef(fx1MARy.MI),
coef(fx1MNAR.MI.u), coef(fx2MNAR.MI.u), coef(fx1MNAR.MI)
),
digits = 3
)
# Table 7.6 #
# Regression coefficients and performqnce of x1+x2 model after MI with aregImpute
print(rbind(
c(coef(fx1MCAR.MI), fx1MCAR.MI$stats["R2"]),
c(coef(fx1MARx.MI), fx1MARx.MI$stats["R2"]),
c(coef(fx1MARy.MI), fx1MARy.MI$stats["R2"]),
c(coef(fx1MNAR.MI), fx1MNAR.MI$stats["R2"])
),
digits = 3
)
## End x, start y ##
g <- aregImpute(~ yMCAR + x1 + x2, n.impute = m, data = dy, pr = F, type = "pmm")
fy.x1.MCAR.MI.u <- fit.mult.impute(yMCAR ~ x1, ols, xtrans = g, data = dy, pr = F)
fy.x2.MCAR.MI.u <- fit.mult.impute(yMCAR ~ x2, ols, xtrans = g, data = dy, pr = F)
fyMCAR.MI <- fit.mult.impute(yMCAR ~ x1 + x2, ols, xtrans = g, data = dy, pr = F)
g <- aregImpute(~ yMARx2 + x1 + x2, n.impute = m, data = dy, pr = F, type = "pmm")
fy.x1.MAR.MI.u <- fit.mult.impute(yMARx2 ~ x1, ols, xtrans = g, data = dy, pr = F)
fy.x2.MAR.MI.u <- fit.mult.impute(yMARx2 ~ x2, ols, xtrans = g, data = dy, pr = F)
fyMAR.MI <- fit.mult.impute(yMARx2 ~ x1 + x2, ols, xtrans = g, data = dy, pr = F) ## areg
g <- aregImpute(~ yMNAR + x1 + x2, n.impute = m, data = dy, pr = F, type = "pmm")
fy.x1.MNAR.MI.u <- fit.mult.impute(yMNAR ~ x1, ols, xtrans = g, data = dy, pr = F)
fy.x2.MNAR.MI.u <- fit.mult.impute(yMNAR ~ x2, ols, xtrans = g, data = dy, pr = F)
fyMNAR.MI <- fit.mult.impute(yMNAR ~ x1 + x2, ols, xtrans = g, data = dy, pr = F) ## areg
# Regression coefficients after MI with aregImpute
print(rbind(
coef(fy.x1.MCAR.MI.u), coef(fy.x2.MCAR.MI.u), coef(fx1MCAR.MI),
coef(fy.x1.MAR.MI.u), coef(fy.x2.MAR.MI.u), coef(fyMCAR.MI),
coef(fy.x1.MNAR.MI.u), coef(fy.x2.MNAR.MI.u), coef(fyMNAR.MI)
),
digits = 3
)
# Regression coefficients and performance after MI with aregImpute
print(rbind(
c(coef(fyMCAR.MI), fyMCAR.MI$stats["R2"]),
c(coef(fyMAR.MI), fyMAR.MI$stats["R2"]),
c(coef(fyMNAR.MI), fyMNAR.MI$stats["R2"])
),
digits = 3
)
#### End Table 7.6 illustration missing values and imputation ####
#### Repeat for binary outcomes ####
library(mice)
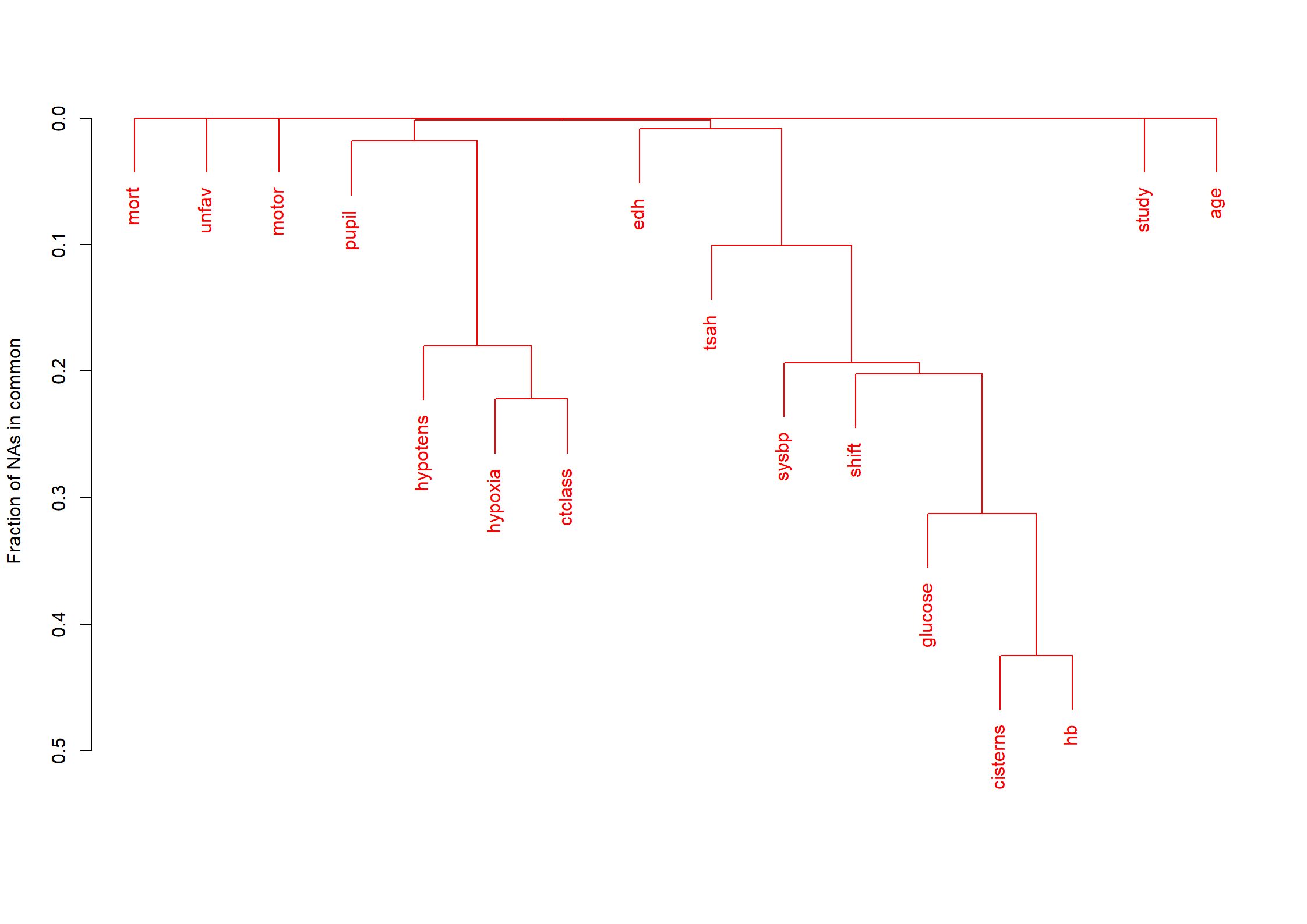
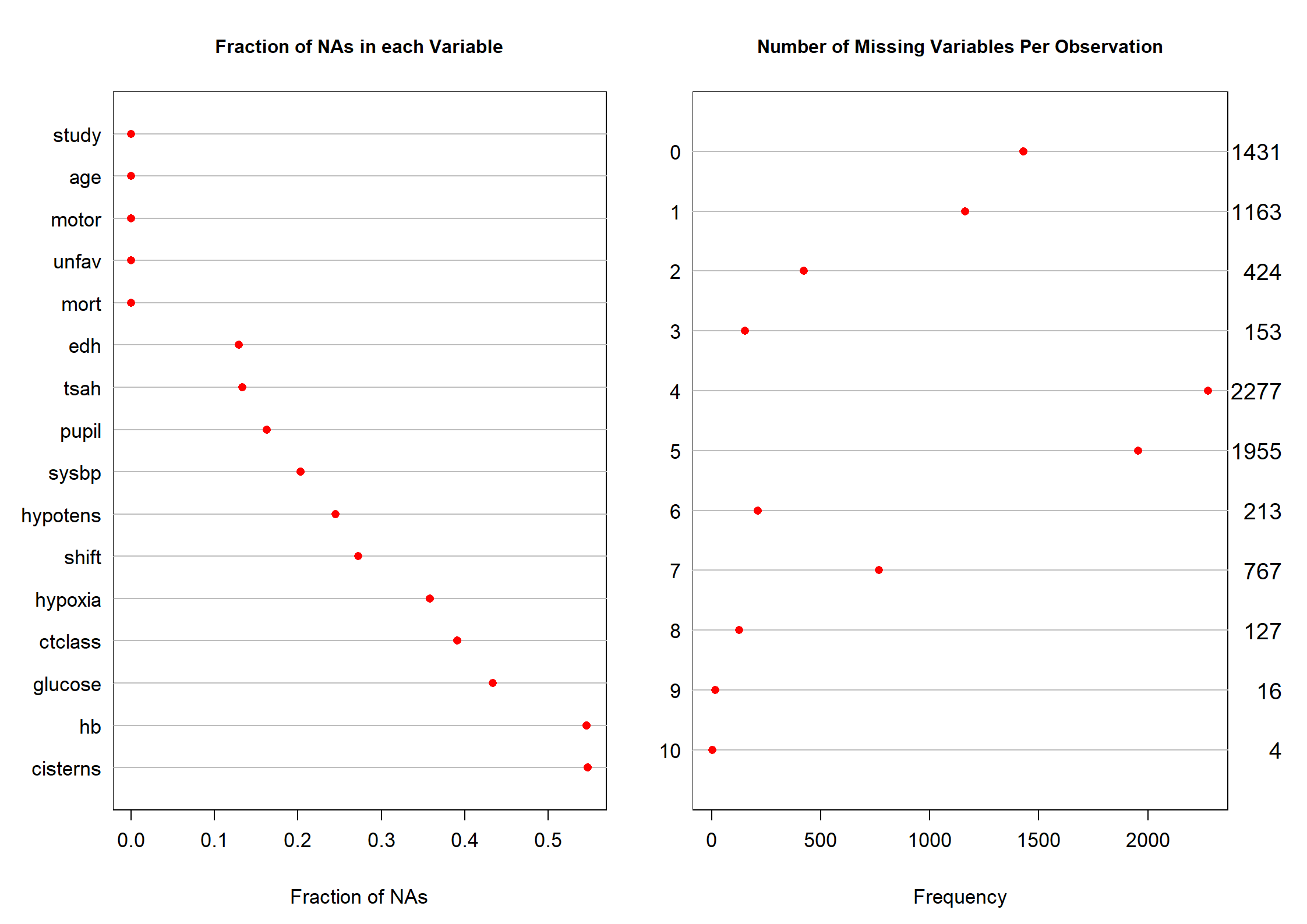
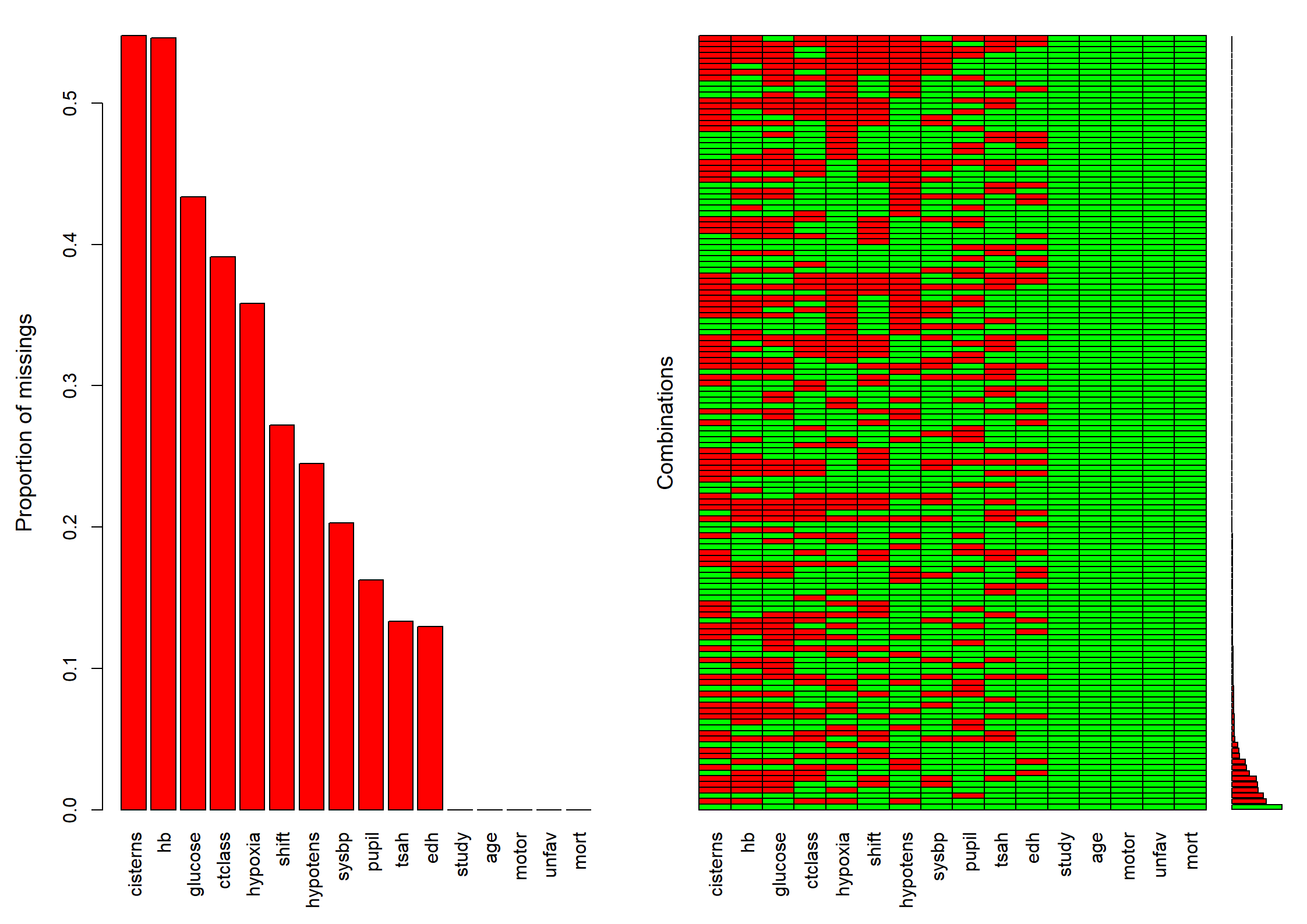
md.pattern(dMNAR)
na.pattern(dMNAR)
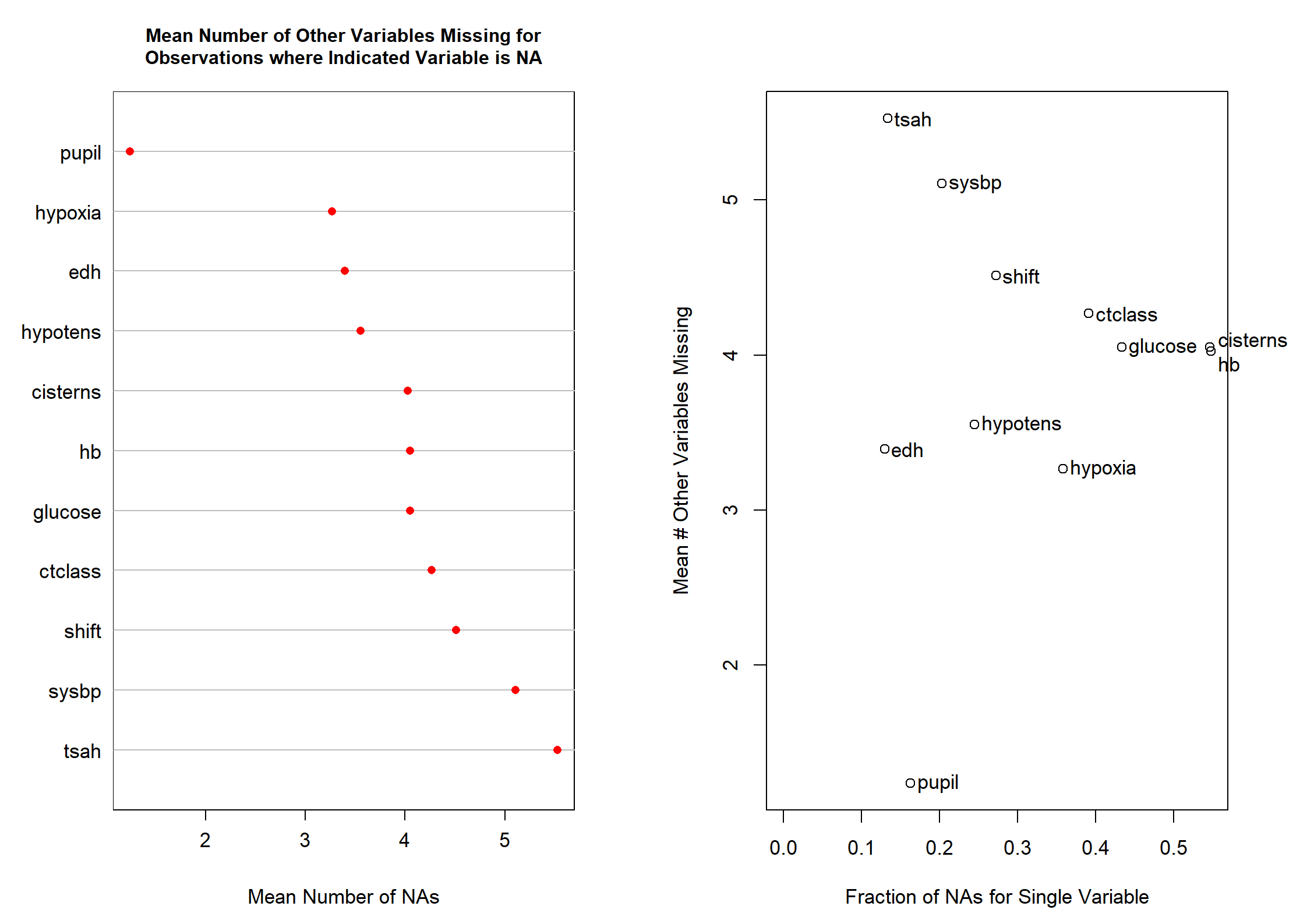
naclus(dMNAR)
set.seed(1) # For identical results at repetition
n <- 100000 # arbitrary sample size
x2 <- rnorm(n = n, mean = 0, sd = 1) # x2 standard normal
x1 <- rnorm(n = n, mean = 0, sd = 1) # Uncorrelated x1
y1.bin <- rbinom(n, 1, plogis(1 * x1 + 1 * x2)) # generate y
x1MCAR <- ifelse(runif(n) < .5, x1, NA) # MCAR mechanism for 50% of x1
x1MARx <- ifelse(rnorm(n = n, sd = .8) < x2, x1, NA) # MAR on x2, R2 50%, 50% missing (since mean x2==0)
x1MARy <- ifelse(rbinom(n, 1, .8 * y1.bin) == 1 | rbinom(n, 1, .2 * (1 - y1.bin)) == 1, NA, x1) # MAR on y, R2 50%, 50% missing (since mean y1==0)
x1MNAR <- ifelse(rnorm(n = n, sd = .8) < x1, x1, NA) # MNAR on x1, R2 50%, 50% missing (since mean x1==0)
mean(is.na(x1MARy)[y1.bin == 0])
mean(is.na(x1MARy)[y1.bin == 1])
lrm(y1.bin ~ x1 + x2)
lrm(y1.bin ~ x1MCAR + x2)
lrm(y1.bin ~ x1MARx + x2)
lrm(y1.bin ~ x1MARy + x2)
lrm(y1.bin ~ x1MNAR + x2)
# Compare fits in the various selections
fx1.CC <- lrm(y1.bin ~ x1 + x2)
fx1MARx.CC <- lrm(y1.bin ~ x1MARx + x2)
fx1MARy.CC <- lrm(y1.bin ~ x1MARy + x2)
fx1MNAR.CC <- lrm(y1.bin ~ x1MNAR + x2)
# Regression coefficients
print(rbind(
coef(fx1.CC), coef(fx1MARx.CC),
coef(fx1MARy.CC), coef(fx1MNAR.CC)
), digits = 3)
print(rbind(
sqrt(diag(fx1.CC$var)), sqrt(diag(fx1MARx.CC$var)),
sqrt(diag(fx1MARy.CC$var)), sqrt(diag(fx1MNAR.CC$var))
), digits = 3)
# Imputation; make data sets with different types of missings
d <- as.data.frame(cbind(y1.bin, x1, x2, x1MCAR, x1MARx, x1MARy, x1MNAR))
dMCAR <- d[, c(1, 3, 4)] # data set with x1 missings according to MCAR
dMARx <- d[, c(1, 3, 5)] # x1 MAR on x2
dMARy <- d[, c(1, 3, 6)] # x1 MAR on y
dMNAR <- d[, c(1, 3, 7)] # x1 MNAR at x1
## MI using the aregImpute function from rms
## help(areImpute) for more info
g <- aregImpute(~ y1.bin + x1MCAR + x2, n.impute = 5, data = d, pr = F, type = "pmm")
## fit models per imputed set and combine results using Rubin's rules
## Use fit.mult.impute() function from rms
fx1MCAR.MI <- fit.mult.impute(y1.bin ~ x1MCAR + x2, lrm, xtrans = g, data = d, pr = F)
g <- aregImpute(~ y1.bin + x1MARx + x2, n.impute = 5, data = d, pr = F, type = "pmm")
fx1MARx.MI <- fit.mult.impute(y1.bin ~ x1MARx + x2, lrm, xtrans = g, data = d, pr = F) ## areg
g <- aregImpute(~ y1.bin + x1MARy + x2, n.impute = 5, data = d, pr = F, type = "pmm")
fx1MARy.MI <- fit.mult.impute(y1.bin ~ x1MARy + x2, lrm, xtrans = g, data = d, pr = F) ## areg
g <- aregImpute(~ y1.bin + x1MNAR + x2, n.impute = 5, data = d, pr = F, type = "pmm")
fx1MNAR.MI <- fit.mult.impute(y1.bin ~ x1MNAR + x2, lrm, xtrans = g, data = d, pr = F) ## areg
# Regression coefficients after MI with aregImpute
print(rbind(
coef(fx1MCAR.MI), coef(fx1MARx.MI),
coef(fx1MARy.MI), coef(fx1MNAR.MI)
), digits = 3)
print(rbind(
sqrt(diag(fx1MCAR.MI$var)), sqrt(diag(fx1MARx.MI$var)),
sqrt(diag(fx1MARy.MI$var)), sqrt(diag(fx1MNAR.MI$var))
), digits = 3)